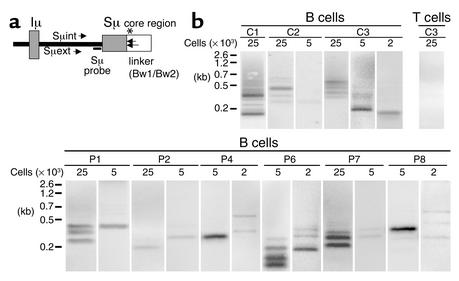
Figure 4.
Detection of double-strand DNA breaks (DSBs) during CSR in HIGM4. (a) Positions of primers and probe used in DSB assay. Directions and positions of the primers and radioactive Sμ-specific probe used in the DSB assay are shown. The asterisk shows the putative position of DSBs in the Sμ region. (b) DSBs were detected by LM-PCR in activated B cells of HIGM4 patients. Genomic DNA of sCD40L+rIL-4–activated B cells (2 × 103, 5 × 103, or 25 × 103 cells per lane) from controls (C1, C2, C3) or HIGM4 patients (n = 6, P1, P2, P4, P6–P8) was ligated with a double-stranded linker and amplified by a semi-nested PCR using the linker (Bw1) as a primer as well as two specific primers in the Sμ region (Sμext and Sμint). The PCR products were subjected to electrophoresis and transferred onto a membrane before being incubated with a radiolabeled Sμ-specific probe. Radioactivity was detected by use of a PhosphorImager. The negative control consisted of anti-CD3+rIL-2–activated T cells (25 × 103 cells per lane, C3).