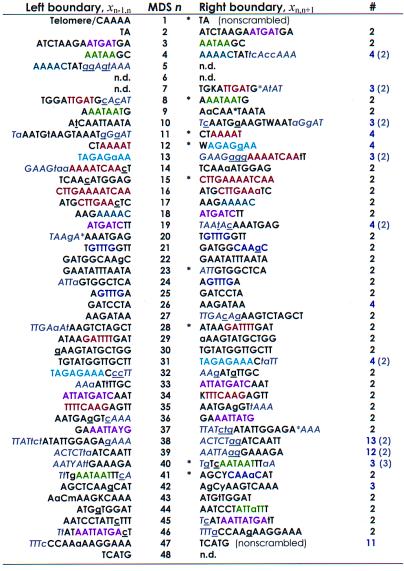
Table 1.
Stylonychia lemnae DNA pol α pointers plus context are unique
This table lists the boundary sequences (pointers xij) at the ends of each MDS in the micronuclear DNA. Flanking sequences in gray and italics provide extended context for pairing. Sequences that are identical between MDS n and n + 1 are in uppercase; asterisks within a sequence represent gaps introduced to maximize alignment (by eye). Where there are mismatches (lowercase), the macronuclear sequence contains the underlined nucleotide. The left boundary of MDS 1 begins with the encoded sequence CAAAA, which serves as a telomere addition site. Pointers marked by an asterisk are the most conserved among S. lemnae, O. nova, and O. trifallax. The right column indicates the multiplicity of each pointer (number of times the identical sequence is found in either strand of the major or minor locus). Numbers in parentheses include an additional gray sequence, which generally reduces the multiplicity of each pointer to two, the expected redundancy at the end of MDS n, and beginning of MDS n + 1. Pointers with multiplicity greater than 2 (even with additional sequences) all follow short (9–28 bp) MDSs that are exchanged for IESs of similar length (MDS 11, 12, 26, 40, 42; Table 2), suggesting that the physical alignment provides the context for pairing. Sequences to the left of MDS 30 and the right of MDS 46 were part of PCR primers and were not sequenced directly. IUPAC symbols specify pairs of nucleotides present at positions that differ between alleles. Sequence motifs (representative “words” or “subwords” that recur on either strand) shared between two or more pointer pairs are indicated by the same color.