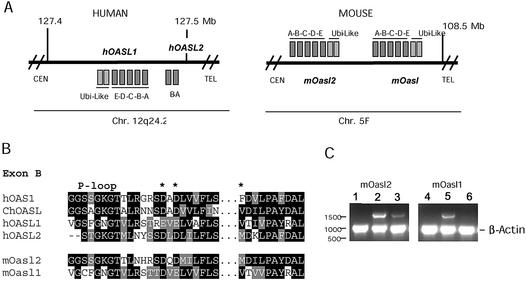
Figure 1.
(A) Schematic drawing of the hOASL and mOasl genes on human chromosome 12 and mouse chromosome 5. Genes with reading frames in the same direction as the annotated nucleotide sequence are indicated by squares above the lines; genes below are in the reverse direction. The genes are not drawn to scale. CEN, centromere; TEL, telomere. (B) Amino acid alignments of hOAS1 p42 (D00068), ChOASL (AB037592), hOASL1 (AJ22089), hOASL2 (NT_028327), mOasl1 (AY089728) and mOasl2 (AK010034) (numbers in parentheses indicate the GenBank accession no. for each protein). Black boxes indicate conserved residues, grey boxes related sequences. Gaps are indicated by –. The middle part of exon B has been cut out as indicated by black dots. The three catalytic aspartate residues are indicated by asterisks. (C) Transcription of mOasl2 (lanes 1–3) and mOasl1 (lanes 4–6) genes. Products of PCRs (30 cycles) with cDNA from untreated (lanes 1 and 4), IFN-α-induced (lanes 2 and 5) and IFN-γ-induced (lanes 3 and 6) mouse L929 cells, analysed by gel electrophoresis (1% agarose). The mOasl reaction and the control reaction (murine β-actin) were mixed in equal amounts before gel electrophoresis.