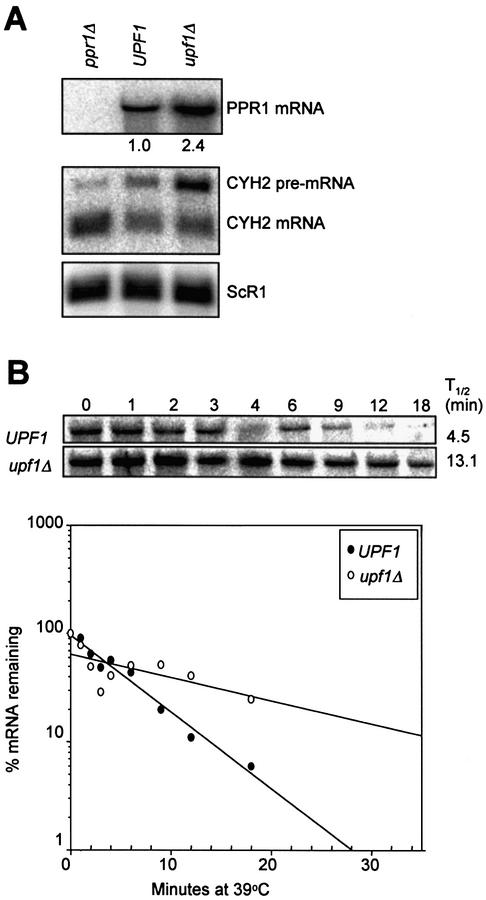
Figure 1.
The decay of wild-type PPR1 mRNA is Upf1p-dependent. (A) PPR1 mRNA accumulation in BY4741 (ppr1Δ) grown in YPD, and PLY167 [pAA79] (UPF1) and PLY167 [pRS315] (upf1Δ) grown in CM-leucine. Northern blots were prepared with 15 µg of total RNA and hybridized with radiolabeled PPR1, CYH2 and ScR1 DNA. CYH2 pre-mRNA is a target for NMD used to confirm the NMD phenotype of our yeast cells (18). ScR1 mRNA is transcribed by RNA polymerase III and is not degraded by NMD (33,34). It is used as a loading control. The relative PPR1 steady-state levels are indicated under the northern blot hybridized with radiolabeled PPR1 DNA. (B) PPR1 mRNA half-lives were measured in isogenic yeast strains AAY334 (UPF1, upper image) and AAY335 (upf1Δ, lower image) both grown in YAPD. The PhosphorImages are of representative northern blots hybridized with radiolabeled PPR1 DNA. The northern blots were prepared with 15 µg total RNA harvested at different time points following inhibition of RNA polymerase II. The percent mRNA remaining at each time point following inhibition of transcription was calculated by dividing the amount of probe hybridized to a particular band (corrected for loading with ScR1) by the amount of probe hybridized to the band at time point zero. The percent mRNA remaining versus time after transcriptional inhibition was plotted using Sigmaplot™. The PPR1 mRNA half-lives was determined and is shown to the right of the PhosphorImages.