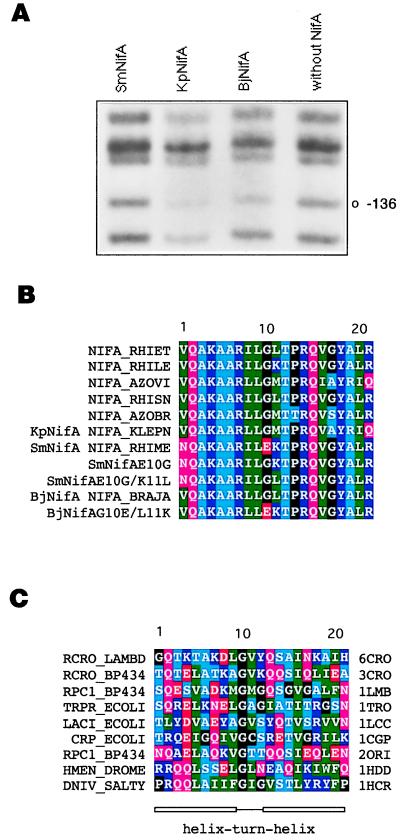
Figure 1.
DNA-binding properties of different NifA proteins and amino acid sequence of various hth motifs. (A) In vivo DMS footprinting of the K. pneumoniae nifH enhancer with different NifA proteins, as indicated. Protection from methylation of guanine-136 by KpNifA and BjNifA is indicated. This residue is part of the TGT-N10-ACA nifH enhancer. (B) Sequence alignment of the hth motif of several NifA and the SmNifA and BjNifA mutant proteins constructed in this work. The SwissProt name for each protein is indicated. (C) Alignment of the hth motifs of proteins whose structures have been solved as cocrystals with their DNA-binding sites. SwissProt (first column) and Protein Data Bank (last column) names are specified. 6CRO, lambda cro; 3CRO, 434 cro; 1LMB, lambda repressor; 1TRO, trp repressor; 1LCC, lac repressor; 1CGP, catabolite activator protein (cap); 2ORI, 434 repressor; 1HDD, Drosophila homeodomain protein; 1HCR, Hin recombinase. A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.