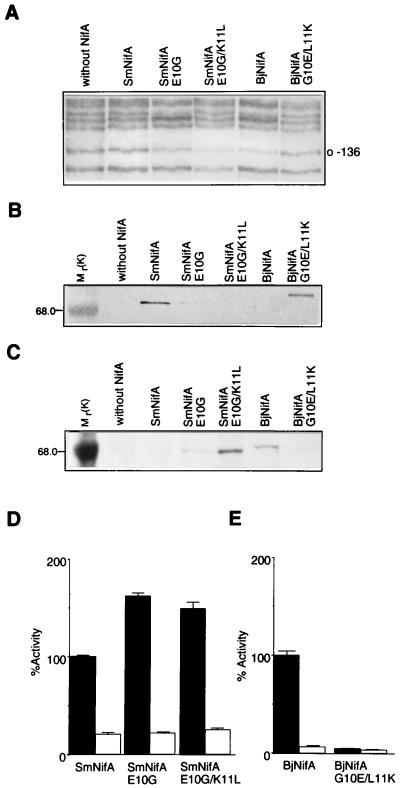
Figure 2.
Analysis of DNA binding, protein stability, and transcriptional activation of SmNifA, BjNifA, and their mutant derivatives. (A) Footprinting in vivo of the K. pneumoniae nifH enhancer with different NifA proteins and mutant derivatives, as indicated. Guanine-136 of the nifH enhancer is marked. (B and C) Immunodetection of the SmNifA and BjNifA (see Materials and Methods) proteins and their mutant derivatives. Antibodies raised against synthetic peptides corresponding to the hth of SmNifA or BjNifA were used for the immunodetection shown in B and C, respectively. Mr values of control proteins are denoted. Note that the anti-SmNifA hth peptide antibodies did not react against either BjNifA or SmNifAE10G/K11L and only weakly against SmNifAE10G, but efficiently recognized BjNifAG10E/L11K. Conversely, antibodies raised against BjNifA hth peptide did not recognize either SmNifA or BjNifAG10E/L11K and reacted weakly against SmNifAE10G, but efficiently recognized SmNifAE10G/K11L. (D and E) Transcriptional activation of a nifH-lacZ fusion carrying (solid bars) or deleted of (open bars) the enhancer by different NifA proteins and their mutant derivatives, as indicated. Because SmNifA and BjNifA are coded in different vectors and expressed from different promoters, it is not feasible to correlate actual β-galactosidase activities. Values represent the mean ± SD of three independent experiments and are expressed as percentage of β-galactosidase activity in Miller units.