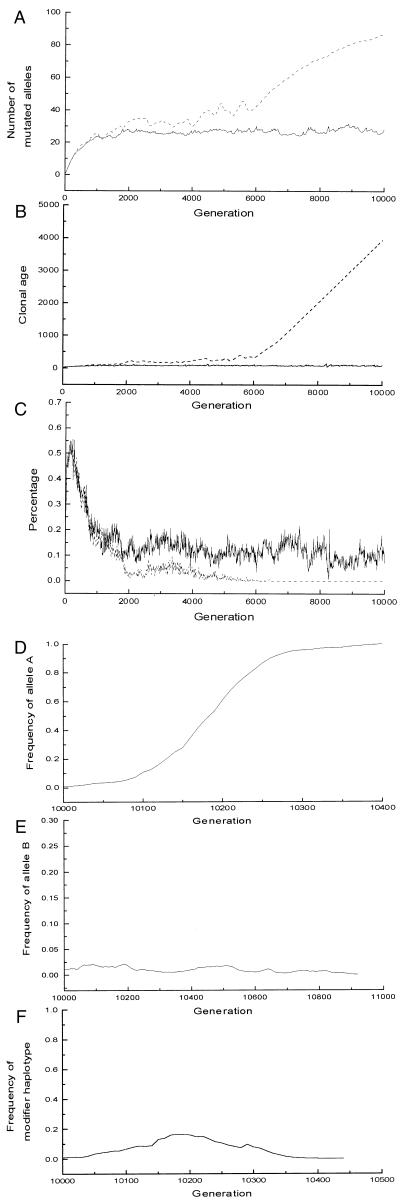
Figure 6.
The results of the individual-based model using a modified fitness function (ps = 0.025, μ = 0.05, s = 0.05, N = 100, AL = 200). The dashed line stands for the species without cell senescence; the solid line stands for the species that employs cell senescence. (A) Average number of deleterious mutations per diploid genome. (B) Average clonal age. (The average clonal age of individuals in the species employing cell senescence fluctuates around 70.) (C) Percentage that offspring from sexual reproduction that have above-average fitness. After 10,000 generations, modifier alleles were introduced into the population. (D) Asexual reproduction modifier allele A invades the population without cell senescence. (The mutant's genotype is supposed to be AB/AB; the initial frequency of allele A is 0.01. If the genotype is AB/aB, the frequency of A will converge to 0.5. Either way the mutants will invade the whole population.) (E) The modifier allele B cannot invade the population with cell senescence (initial frequency of allele B is 0.01). (F) Frequency of modifier haplotypes AB in the population with cell senescence. (Here we suppose the genotype of mutants is AB/ab. Similar result can be produced by using other genotypes, e.g., AB/AB.)