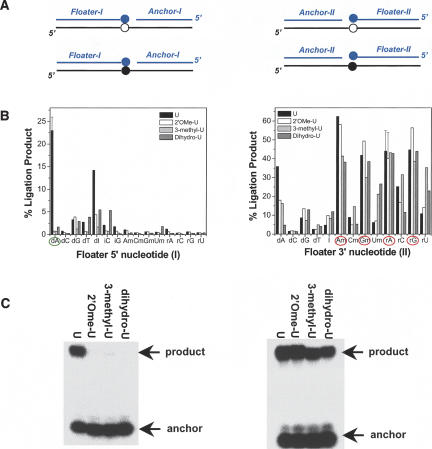
FIGURE 2.
Finding D- and N-oligos for each modification type. (A) Two sets of oligo pairs with the ligation junction 5′ (set I) or 3′ (set II) to the modification site. Black = model 30-mer RNA containing unmodified (open) or modified (filled circles) nucleotide. Blue = floater and anchor oligonucleotides as ligation substrates. (B) A total of 15 set I pairs and 13 set II pairs are tested for 2′O-methyl-U, 3-methyl-U and 5,6-dihydro-U modifications. I-dA is the only sufficiently discriminating D-oligo among these 28 oligo pairs, whereas II-rA, II-Am, II-rG, and II-Gm can be used as N-oligos. (C) Ligation using the designated D-oligo (I-dA floater) to discriminate unmodified U from 2′Ome-U, 3-methyl-U and 5,6-dihydro-U (left, >10-fold). Ligation using a designated N-oligo (II-rA floater) shows <1.3-fold variation between the unmodified U and the other modifications (right).