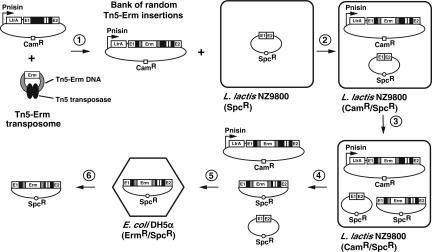
FIGURE 3.
Genetic screen to identify potential insertion sites within Ll.LtrB. In the presence of the Tn5 transposase (black ovals), the Tn5-Erm transposon DNA (Tn5-Erm DNA) was mixed in a 1:1 molar ratio with a plasmid harboring the Ll.LtrB construct that expresses LtrA in trans (step 1). This in vitro reaction created a saturated bank of random Tn5 insertions around the donor plasmid, consisting of insertions between every nucleotide within the intron. This bank of random Tn5-Erm insertions was transformed in NZ9800 L. lactis cells, which contained the recipient plasmid pMNHS (step 2). Intron expression was induced with nisin (step 3), the plasmid mix (donor plasmid, recipient plasmid, and mobility products) was recovered (step 4), and then retransformed in E. coli DH5α cells (step 5). E. coli cells were plated on LB/Erm/Spc plates to select for Ll.LtrB mobility products that carried a copy of the Tn5-Erm transposon. Only introns that were still mobile following the insertion of a Tn5-Erm transposon could invade the recipient plasmid. Twenty independent Ll.LtrB mobility products were recuperated (step 6), and the locations of Tn5 insertions were analyzed by sequencing the intron–transposon junctions from both ends of the transposon. This screen was repeated with a Tn5-Kan transposon.