Abstract
A molecular understanding of synaptogenesis is a critical step toward the goal of understanding how brains “wire themselves up,” and then “rewire” during development and experience. Recent genomic and molecular advances have made it possible to study synaptogenesis on a genomic scale. Here, we describe the results of a screen for genes involved in formation and development of the glutamatergic Drosophila neuromuscular junction (NMJ). We screened 2185 P-element transposon mutants representing insertions in ≈16% of the entire Drosophila genome. We first identified recessive lethal mutants, based on the hypothesis that mutations causing severe disruptions in synaptogenesis are likely to be lethal. Two hundred twenty (10%) of all insertions were homozygous lethal. Two hundred five (93%) of these lethal mutants developed at least through late embryogenesis and formed neuromusculature. We examined embryonic/larval NMJs in 202 of these homozygous mutants using immunocytochemistry and confocal microscopy. We identified and classified 88 mutants with altered NMJ morphology. Insertion loci in these mutants encode several different types of proteins, including ATP- and GTPases, cytoskeletal regulators, cell adhesion molecules, kinases, phosphatases, RNA regulators, regulators of protein formation, transcription factors, and transporters. Thirteen percent of insertions are in genes that encode proteins of novel or unknown function. Complementation tests and RT-PCR assays suggest that approximately 51% of the insertion lines carry background mutations. Our results reveal that synaptogenesis requires the coordinated action of many different types of proteins—perhaps as much as 44% of the entire genome—and that transposon mutageneses carry important caveats that must be respected when interpreting results generated using this method.
Keywords: Drosophila, synapse development, axon guidance, axon branching, neuromuscular junction
INTRODUCTION
Neuronal communication depends critically upon the formation of synaptic connections. The human brain has been estimated to contain 1011 neurons, each of which may make and receive hundreds of synaptic contacts. For efficient brain function, it is imperative that each neuron find its cognate partners and form synapses at the appropriate location. Once these initial connections between neurons are made, the structure and functional properties of synapses can be remodeled during development and throughout the lifespan of the organism. Furthermore, many forms of synaptic plasticity, such as use-dependent synaptic strengthening underlying learning and memory, are thought to involve the same mechanisms employed during synapse development (Goda and Davis, 2003; Nordeen and Nordeen, 2004). Not unexpectedly, therefore, the mechanisms controlling synaptogenesis are of intense interest. For over a century, a large amount of neuroscientific effort has been directed toward an understanding of how neurons form and remodel connections. Through this work, a great many insights have been gained, but a clear molecular understanding of synaptogenesis remains elusive.
Of all the techniques developed for the identification and study of individual proteins involved in a biological process such as synaptogenesis, genetics is arguably one of the most powerful. Genetic screens and directed mutation of candidate genes have revealed many important aspects of synapse development (Hortsch and Goodman, 1991; Broadie, 1998; Sanes et al., 1998; Ackley and Jin, 2004). In general, these analyses have focused on specific proteins and the role of each protein in a particular aspect of synapse formation. However, recent genomic and molecular advances now make it possible to address equally important large-scale questions, such as: What percentage of the genome is required for synaptogenesis? Does synaptogenesis rely on any particular types of proteins and how many different types of protein are required? At the molecular level, how similar might axon outgrowth and synapse formation be to other forms of plasma membrane protrusion, cell-cell adhesion, and cell junction maintenance? Answers to these questions are critical for understanding whether synaptogenesis is a unique cellular process or comparable to other cell growth processes and cell-cell interactions.
Here, we describe the results of a genome-wide screen for transposon insertion mutants affecting formation of Drosophila neuromuscular junctions (NMJs). The Drosophila NMJ is a relatively well-described and experimentally accessible model glutamatergic synapse that has been widely used for the study of synapse development and function. A large part of this model synapse’s attraction is the fact that many molecular and genetic tools are available for use in Drosophila. One important tool, that we rely on here, is provided by the BDGP Gene Disruption Project. The Gene Disruption Project has so far has generated P-element insertions in approximately half of all Drosophila genes (Bellen et al., 2004). The mutants generated by this project, in combination with those developed by others, will most likely eventually result in transposon insertions in almost all Drosophila genes within the next few years. Transposon mutants are particularly inviting mutants because a transposon insertion effectively “tags” the insertion locus, making identification of the mutant gene relatively straightforward. Previous forward genetic screens for fly NMJ mutants have successfully identified important synaptogenesis proteins (Kraut et al., 2001; Parnas et al., 2001; Aberle et al., 2002; McCabe et al., 2004; Yeh et al., 2005), but these screens typically used chemical mutagens, which are effective but require subsequent laborious mapping to identify the mutant gene. This limits the ability to perform genome-scale analyses. Ours is not the first screen of transposon insertions for fly NMJ mutants. Previous P-element transposon insertions have successfully identified dozens of genes important for regulation of Drosophila NMJ formation (Kraut et al., 2001; Laviolette et al., 2005). However, the “EP”elements used in these studies were engineered to cause gene overexpression, such that the screen identified only gain-of-function mutants. An equivalent loss-of-function screen (such as we describe here) has not been reported. Analysis of the BDGDP transposon mutants also allows us to address important methodological questions. Transposon mutagenesis and “gene-tagging” are increasing lauded, particularly in mammals, as important breakthroughs for genetic modification (Bestor, 2005; Collier et al., 2005; Dupuy et al., 2005). But how reliable is transposon-mediated mutagenesis and gene-tagging for identification of specific proteins involved in a process? Our results highlight important genetic caveats that need to be respected as use of this technology becomes more widespread.
MATERIALS AND METHODS
Lethal GT1 and SuPor-P P-element insertions were identified using the FlyBase Insertions Query Form (http://flybase.bio.indiana.edu/). Stocks containing lethal P-inserts were then obtained from the Bloomington Stock Center (http://flystocks.bio.indiana.edu/). To examine and/or collect embryos and larvae for experiments, GFP-balanced P-element stocks were allowed to lay eggs on agar plates for 3 h at 258C. Plates were then incubated at 258C and periodically examined to determine the latest lethal mutant stage (the age at which no non-GFP animals survived). Once this stage was determined, experiments were performed on animals at the latest viable stage. P-element inserts were rebalanced using appropriate GFP balancers and homozygous mutants on the plate were identified by lack of fluorescence from a GFP balancer under an Olympus SZX12 binocular microscope equipped with epifluorescence and appropriate filters for GFP. Control genotypes used in this study were w1118 or heterozygous siblings from the same laying plate.
Complementation analysis was performed by crossing GFP-balanced P-element mutant stocks to a balanced stock containing a deficiency that removes the insertion site, or a lethal mutation in the gene carrying the insertion. The F1 generation of each cross was examined for the presence or absence of adult flies carrying neither balancer chromosome. Thus, P-element insertion chromosomes were tested for their ability to complement, with regard to viability, lethal mutations in the gene carrying the P-element insertion.
For immunocytochemistry, larvae were manually dissected and fixed for 30-60 min in Bouin’s fixative. Late stage embryos were dechorionated in bleach and then manually devitellinated and dissected, as previously described (Featherstone et al., 2000). All dissections were performed in Roger’s Ringer solution (135 mM NaCl, 5 mM KCl, 4mM MgCl, 1.8 mM CaCl, 5 mM TES, 72 mM sucrose) at room temperature. To visualize NMJs, fluorescently conjugated anti-HRP (Jackson ImmunoResearch Labs, West Grove, PA) was used at 1:100. Confocal images were obtained using an Olympus FV500 laser-scanning confocal microscope.
Semiquantitative RT-PCR for evaluating whether insertions affected transcript size and/or abundance was performed as follows: total RNA was isolated from homozygous mutant embryos (≈50 embryos/reaction) or heterozygous sibling controls using homogenization and trizol extraction (Roberts, 1998). RNA was then reverse transcribed (Superscript II reverse transcriptase; Invitrogen) into cDNA using gene-specific primers and standard methods. Select cDNAs were then PCR-amplified using transcript-specific primers, separated electrophoretically, and visualized using ethidium bromide.
Real time RT-PCR of glutamate receptor subunits in pathfinding/target recognition mutants (Fig. 2) was performed as previously described (Featherstone et al., 2005).
Figure 2.
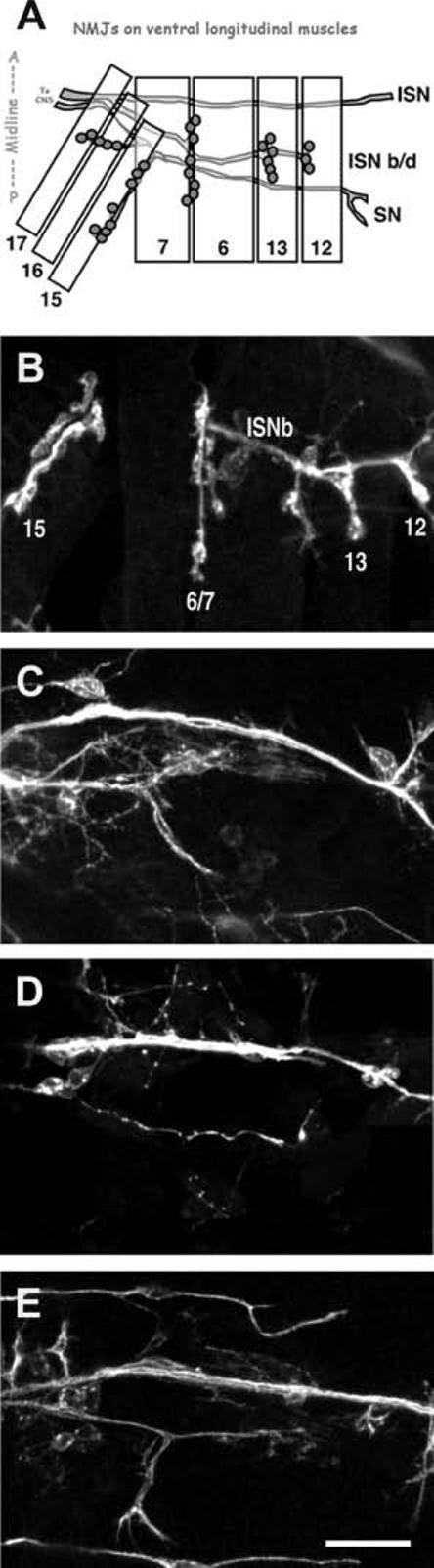
Pathfinding/target recognition phenotypes observed in the screen. (A) Schematic showing wild-type neuromuscular junctions on the ventral body wall muscles in one hemisegment of a Drosophila embryo. Muscles are depicted as numbered rectangles. Intersegmental nerve (ISN) branch b/d (ISNb/d) branches to form NMJs on muscles 7, 6, 13, and 12. The segmental nerve (SN) innervates muscles 17, 16, and 15. NMJ presynaptic arborizations are comprised of several clustered boutons (shaded circles). (B) Confocal image of control embryonic neuromuscular junctions, as diagrammed in (A), visualized using anti-HRP antibodies that stain all neuronal membrane. (C-E) Confocal images of pathfinding/target finding mutants, which do not form neuromuscular junctions on the body wall muscles. (C) w1118; P{GT1}SmBBG02775; (D) y1w67c23; P{SuPor-P}KG02164; (E) y1; P{SuPor-P}KG04415. Scale bar = 10 ≈m.
RESULTS
Screen
To identify genes required for the proper formation of synapses, we screened lethal P-element transposon insertion mutants generated by the BDGP Gene Disruption Project (Bellen et al., 2004) for defects in the presynaptic morphology of Drosophila embryonic and larval 6/7 NMJs. This screen involved several steps (Fig. 1).
Figure 1.
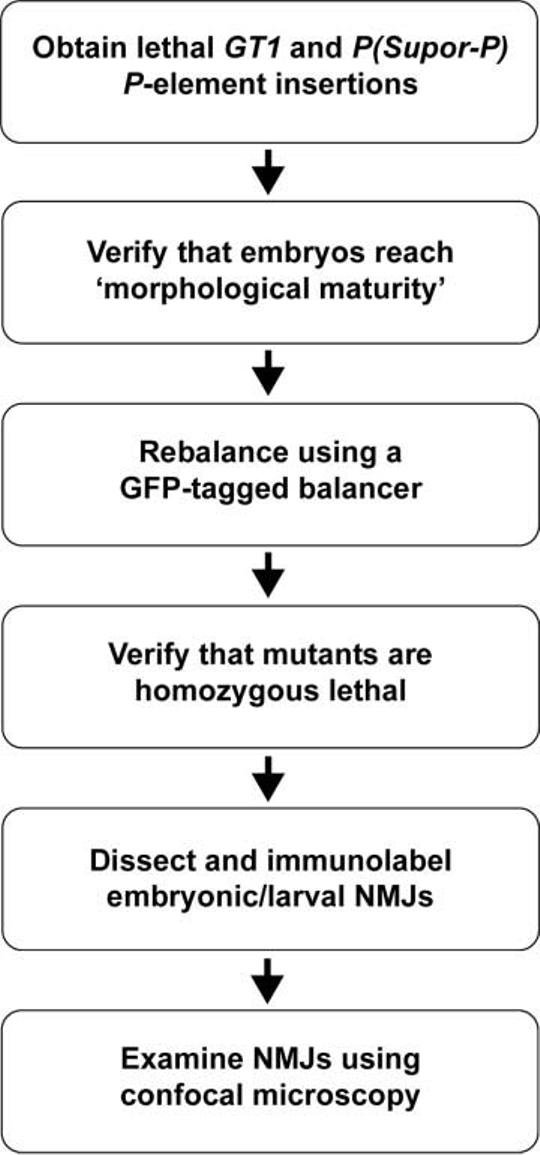
Flow chart outlining the process used to isolate P-element insertion mutants with defects in NMJ development.
We first identified all GT1 and SuPor-P transposon insertion mutants available in March 2003 using the FlyBase Insertions query form (http://flybase.bio.indiana.edu/transposons/fbinsquery.hform). We focused on GT1 and SuPor-P insertion lines because these P-elements are designed to maximally disrupt genes (Roseman et al., 1995; Lukacsovich et al., 2001; Bellen et al., 2004). The severe defects in NMJ formation that we sought to identify commonly cause lethality. Therefore, we identified, using the FlyBase insertions query form, all of the lethal GT1 and SuPor-P insertion mutants that were annotated as “not viable” or requiring a balancer chromosome for stock maintenance. Of the 2185 total mutants considered (predicted to mutate approximately 16% of the entire genome), 220 (≈10%) of the GT1 and SuPor-P insertion lines contained lethal P-element insertions (Table 1).
Table 1.
Lethal Inserts in P-Element Mutants
| Insertion Type | GT1 | P(Supor-P) |
|---|---|---|
| Number of lines | 532 | 1653 |
| Number lethal | 53 | 167 |
| Percent lethal | 10.0 | 10.1 |
A prerequisite for identification of NMJ defects is the formation of neuromusculature. Therefore, we next eliminated from consideration all lethal mutants that were unable to form “morphologically mature” embryos 22-24 h after egg laying (AEL) at 258C. Morphological maturity was evaluated by the presence of typical late stage 17 characteristics such as clear segmentation, mouthhook development, condensation of the CNS, malphigian tubules formation, and visible trachea. Two hundred five of two hundred twenty (93%) of the homozygous lethal mutants formed morphologically mature embryos. This surprisingly high number may reflect a high frequency of genes with maternally contributed RNA and/or protein, rather than a low frequency of genes required for embryonic development.
The next stage of our screen required the unambiguous identification of homozygous mutant embryos and larvae. Therefore, all 205 lethal mutants able to form morphologically mature embryos were rebalanced using an appropriate GFP-tagged balancer chromosome. Absence of green fluorescence allowed the unambiguous selection of homozygous embryos and/or larvae. We were unable to rebalance three mutant lines using chromosome-appropriate GFP-tagged balancers, possibly because the insertion mutant chromosome and balancer chromosome both contained lethal mutations that failed to complement. All 202 GFP-balanced stocks were then checked for the absence of GFP-positive adults. No (0%) GFP-positive adults were found, suggesting that all 202 insertion chromosomes annotated as lethal did indeed carry recessive lethal mutations, at least initially (but see below). We examined F1 offspring every 24 h to determine the latest stage to which homozygous mutant embryos and larvae were viable (Table 2). Of the 202 mutant lines, 22 mutants (11%) failed to hatch, dying as embryos (22-24 h AEL). Fifty-six mutants (27%) died as first instar larvae (<≈ 46 h AEL). Fifty-two mutants (25%) died in either the ≈second (<≈ ≈70 h AEL) or third larval instar (<≈ 120 h AEL). Seventy-two mutants (35%) died during ≈pupation and were unable to eclose.
Table 2.
Homozygous Lethal Stages of P-Element Mutants
| Do Not Hatch | 1stInstar | 2nd/3rdInstar | Pupal | |
|---|---|---|---|---|
| Number of lines | 22 | 56 | 52 | 72 |
| Percentage | 10.7 | 27.3 | 25.4 | 35.1 |
After the lethal stage was determined for each mutant, homozygous animals were manually dissected at the latest viable stage to reveal the NMJs present on ventral longitudinal muscles. Embryonic lethal mutants were dissected and stained as embryos, first instar viable mutants were dissected and stained as first instar larvae, and so on with second and third instar viable mutants. Pupal lethal mutants were dissected and stained as third instar larvae. These preparations were fixed and stained using anti-HRP antibodies, which recognize all Drosophila neuronal membranes, including axons and presynaptic NMJ terminals, and examined for qualitative changes in NMJ development. Specifically, we examined NMJs on ventral longitudinal muscles 6 and 7, which are morphologically distinct, relatively invariant in wildtype, and well described throughout embryonic and larval development (Gramates and Budnik, 1999; Featherstone and Broadie, 2004). NMJs from each of five to ten homozygous animals of each genotype were examined. In many cases, heterozygous siblings were dissected, stained, and examined at the same time. Eighty-eight of the two hundred two rebalanced lethal mutants (44%) displayed qualitative defects in 6/7 NMJ formation and/or development.
The large number of insertion mutants identified in our study (88) suggests that a significant fraction of the genome is required for normal synaptogenesis. The exact fraction depends on whether the exclusion of viable mutants significantly enriched for mutants with synaptogenesis defects. Assuming no enrichment, then our results suggest that approximately 44% (88/202) of the genome is required for normal NMJ development. If every synaptogenesis gene is also lethal when mutated (e.g., perfect enrichment), then approximately 4% (88/2185) of the entire genome is required for normal NMJ development. However, perfect enrichment is unlikely because mild defects in NMJ development are not lethal and not all lethal mutations cause alterations in NMJ development.
Mutant Classification
NMJ development can be broadly separated into two phases: (1) axon pathfinding/target recognition, and (2) synapse differentiation and growth.
Axon pathfinding and target recognition represent the initial stages of NMJ development. Beginning at stage 12 in Drosophila embryonic development, motor neuron growth cones exit the CNS and travel along the developing segmental nerve toward their body wall muscle targets. Approximately 13 h AEL, development of ventral NMJs begins when motor neuron growth cone filopodia contact target muscles. Over the next several hours, filopodia-ringed growth cones in contact with appropriate targets collapse to form nascent presynaptic processes of indistinct shape, termed prevaricosities.
Differentiation and subsequent growth of the presynaptic NMJ arbor represents the second phase of NMJ development. Seventeen to twenty-two hours AEL, parts of the prevaricosities begin to constrict and/or swell such that presynaptic terminals (known as synaptic boutons) are formed (Johansen et al., 1989; Halpern et al., 1991; Sink and Whitington, 1991; Yoshihara et al., 1997; Gramates and Budnik, 1999; Rose and Chiba, 1999; Featherstone and Broadie, 2004). At the time of hatching (22-24 h AEL), presynaptic boutons are still relatively indistinct. During larval development, (24-120 h AEL), the presynaptic arborization grows dramatically in order to accommodate rapidly growing larval muscles. This growth involves additions in NMJ length and bouton number (from about 8-10 in at hatching to 50-100 in the third instar 6/7 NMJ). Boutons also become more distinct during NMJ development, such that the third instar NMJ typically appears as “beads on a string” (Keshishian et al., 1993; Yoshihara et al., 1997; Gramates and Budnik, 1999; Featherstone and Broadie, 2004).
We separated the NMJ phenotypes identified in our study into two groups: 1) mutants with defects in axon pathfinding/target recognition, and 2) mutants with defects in synapse differentiation and growth. The latter group, mutants with defects in synapse differentiation and growth, were further divided into two subgroups: (a) overgrowth mutants (excess boutons and/or branches), and (b) undergrowth mutants (reduced numbers of boutons and/or branches). Thus, all mutants were separated into one of the following three mutually exclusive phenotypic groups: (1) pathfinding/target recognition, (2) overgrowth, or (3) undergrowth (Table 3).
Table 3.
Synaptic Phenotypes of P-Element Mutants
| Pathfinding/ Target Recognition | Overgrowth: More Boutons or Branches | Undergrowth: Fewer Boutons or Branches | |
|---|---|---|---|
| Number of lines | 13 | 47 | 23 |
| Percentage | 16% | 56% | 28% |
Pathfinding/Target Recognition Mutants
In Drosophila embryos and larvae, NMJ morphology is highly stereotyped. Figure 2(A) diagrams NMJs on the ventral longitudinal body wall muscles of a single embryonic hemisegment. Ventral longitudinal muscles 15, 16, 17, 7, 6, 13, and 12 are innervated by various branches of the segmental nerve (SN) and intersegmental nerve (ISN). Axons from ISN branch b/d innervate muscles 6 and 7 along the middle third of the cleft between the two muscle cells, then continue distally to innervate muscles 13 and 12. Figure 2(B) shows a confocal Z-projection of the wild-type ventral longitudinal NMJs, as diagrammed in Figure 2(A). Mutant embryonic NMJs were compared to this highly stereotyped pattern. If embryonic mutant ISNs did not appear to form NMJs, but rather failed to enter the body wall muscle field or bypass the muscles, the mutant was classified as having a defect in pathfinding/target recognition. Figure 2(C-E) shows several examples of ventral longitudinal body wall muscle fields stained with anti-HRP to visualize all neuronal membranes from pathfinding/target recognition mutant embryos. In each case, nerves are present but few, if any, obvious NMJs are formed. Subsequent real-time RT-PCR experiments (data not shown) verified that postsynaptic muscles in the three mutants shown in Figure 2 did not show innervation-induced increases in glutamate receptor transcription, consistent with the conclusion that NMJ formation in these mutants does not initiate. Fourteen of eighty-eight mutants (16%) were classified as having pathfinding/target recognition defects (Table 3). All of the mutants in this category exhibited normal outgrowth of the ISN. In some mutants, the ISN did not branch [as shown in Fig. 2(D)], while in others the ISN appeared to branch normally but failed to innervate the underlying musculature [as shown in Fig. 2(E)]. All pathfinding/target recognition mutants were embryonic or early first instar larval lethal.
Given that leading theories of axon pathfinding and target recognition require multiple guidance and recognition cues, we were surprised that only a minority of our mutants fell into the axon pathfinding/target recognition category. Multiple cues encoded by many different genes should be “hit” with a relatively high frequency in a screen such as ours. Assuming that axon guidance and target recognition genes are not somehow refractory to transposon mutageneses, one possibility is that most pathfinding/target recognition mutants are not lethal, and therefore would not have been identified by our screen (which eliminated from consideration all viable mutants). But this is argued against by the fact that all (100%) of our pathfinding/target recognition mutants were embryonic lethal, suggesting a reasonably strong association between this type of mutant and lethality. Another possibility is that genes required for pathfinding and target recognition are also required for formation of neuromusculature, and thus would be eliminated from consideration early in our screen. However, only 7% of lethal mutants did not form grossly normal embryos with neuromusculature. Therefore, the low recovery of axon pathfinding and target recognition mutants in our screen probably reflects a low incidence of axon pathfinding and target recognition genes in the genome.
Overgrowth Mutants
Mutants with NMJs that displayed an abnormally large number of boutons or branches were classified as “overgrowth mutants.” Figure 3(A-D) shows examples of embryonic/first instar NMJs on ventral longitudinal muscles. Figure 3(A) shows NMJs from control embryos, as diagrammed in Figure 2(A) and replicated in Figure 2(B). Figure 3(B-D) shows homologous NMJs in three different embryonic/L1 lethal overgrowth mutants. Figure 3(E) shows an example of control NMJs on ventral longitudinal muscles 6 and 7 of third instar larvae. Figure 3(F-H) shows homologous muscle 6/7 NMJs in three different larval/pupal lethal overgrowth mutants. In most cases [e.g., Fig. 3(B,F-H)], presynaptic terminal arborizations in overgrowth mutants were abnormally long and/or covered an abnormally large area of postsynaptic muscle. In some cases, however, NMJs with increased numbers of boutons or branches actually extended across a smaller region of the postsynaptic muscle [e.g., Fig. 3(C)]; these NMJs were nonetheless still classified as overgrowth mutants, due to the increased bouton and branch number. One advantage of this classification scheme (rather than one depending on NMJ arborization area) is that it is largely independent of muscle size, and thus largely independent of contamination by myoblast fusion defects or other alterations in muscle growth. Overgrowth mutants were the largest class of mutants identified in this study; 48 of 88 mutants (55%) displayed overgrown NMJs (Table 3). Overgrowth mutants died at varying stages of development, from embryos to pupae. We did not systematically determine whether NMJ morphology was normal at earlier (embryonic-L2) stages in larval lethal NMJ overgrowth mutants; however, this was the case in at least one mutant, P{SuPor-P}CG2095KG02723, studied in more detail (Liebl et al., 2005).
Figure 3.
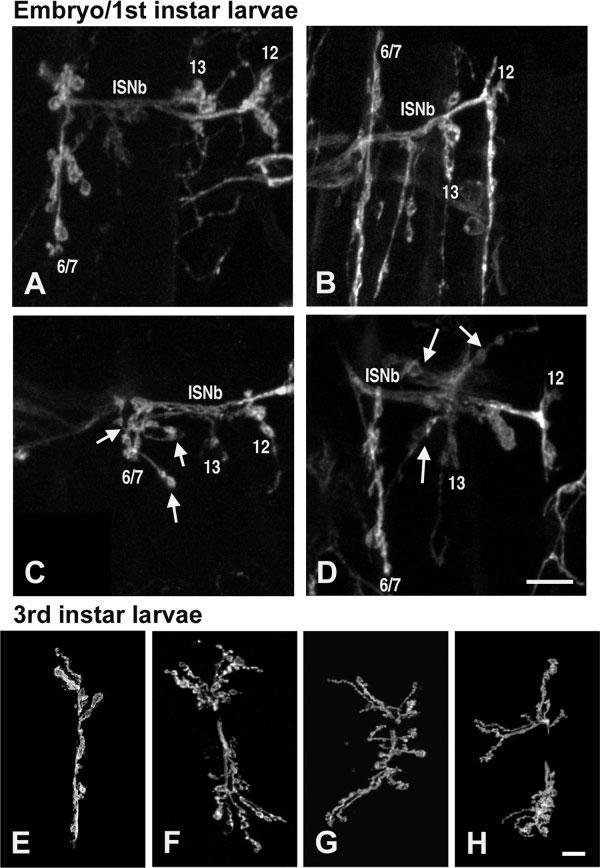
Overgrowth phenotypes observed in the screen. (A-D) Embryonic and first instar larval neuromuscular junctions labeled with the neuronal marker anti-HRP. Scale bar 10 ≈m. (A) Control first instar larval neuromuscular junctions, as in Figure 2(B). (B) y1w67c23; = P{SuPor-P}KG00158 mutants display abnormally long presynaptic arborizations. (C) Muscle 6/7 NMJs in y1; P{SuPor-P}KG01736 mutants exhibit additional branches (arrows) and abnormal morphology. (D) NMJs in y1; ry506 P{SuPor-P}IP3KIKG02192 mutants exhibit additional branches (arrows) and boutons. (E-H) Third instar larval 6/7 neuromuscular junctions labeled with anti-HRP. Scale bar 20 ≈m. (E) Control third instar 6/7 neuromuscular junction. (F-G) Overgrowth mutant third instar larval = neuromuscular junctions have more branches and presynaptic boutons, compared with controls. (F) y1; P{SuPor-P}nufKG02305 ry506;(G) y1; P{SuPor-P}CG15266KG01622;(H) y1; P{SuPor-P}CG8552KG03914.
The surprisingly large number of overgrowth mutants identified in our screen suggests that NMJ growth is actively limited by multiple molecular mechanisms—normal NMJ morphology therefore likely results from a steady-state “balance” between opposing forces of presynaptic growth and limitation.
Unadergrowth Mutants
Homozygous mutants with NMJs that contained fewer branches and/or boutons compared to controls were categorized as “undergrowth” mutants. Mutants that showed qualitatively immature NMJ morphology at time of hatching (22-24 h AEL), possibly representing delayed synaptogenesis, were also classified as undergrowth mutants. For example, the NMJs shown in Figure 4(B) were classified as undergrown due to the lack of distinct boutons; in this mutant, NMJs at normal time of hatching (22-24 h AEL) maintained an appearance more typical of the “prevaricosity” stage (13-17 h AEL). At older stages, undergrowth mutants always displayed obviously contracted presynaptic arborizations covering a smaller than normal fraction of the postsynaptic muscle surface [Fig. 4(D-F)]. Twenty-six of eighty-eight mutants (30%) showed NMJ undergrowth (Table 3). As with overgrowth mutants, undergrowth mutants died at varying stages of development, from embryos to pupae.
Figure 4.
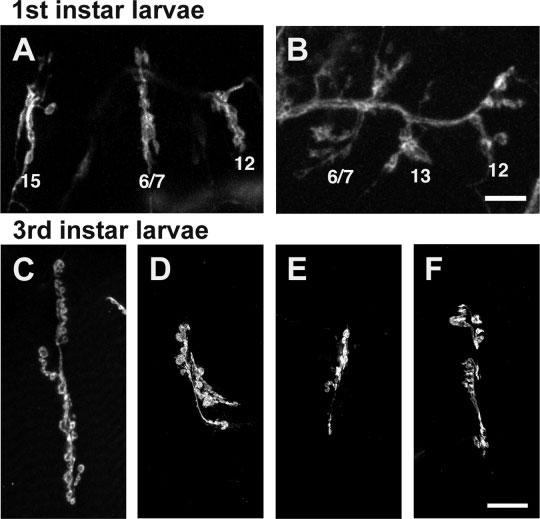
Undergrowth phenotypes observed in the screen. (A,B) Embryonic neuromuscular junctions labeled with anti-HRP. Scale bar = 10 ≈m. (A) Control embryonic neuromuscular junctions, as in Figures 2(B) and 3(A). (B) NMJs in w1118; P{GT1}ftz-flBG01734 mutants display immature morphology. (C-F) Third instar larval 6/7 neuromuscular junctions labeled with anti-HRP. Scale bar = 20 ≈m. (C) Control third instar 6/7 neuromuscular junction, as in Figure 3(E). (D-F) Undergrowth mutant third instar 6/7 neuromuscular junctions exhibit fewer presynaptic boutons compared with controls. (D) w1118; P{GT1}BG01610; (E) y1; P{SuPor-P}KG03611 ry506; (F) y1; P{SuPor-P}RpL3KG05440 ry506.
Probable Function of Insertion Loci
The P-element insertion locations in all of the lethal mutants screened in this study have been determined by inverse PCR, such that each insertion is associated with a specific gene (Bellen et al., 2004). For each of the mutants identified in our screen, we used the BDGDP P-screen database (http://flypush.imgen.bcm.tmc.edu/pscreen/) and/or BLAST searches with flanking genomic sequence from iPCR to identify the genes mutated by the inserted P-element. Putative functions were assigned to each of these genes based on previous publications, FlyBase annotations, and/or GENBANK BLAST searches. We classified the genes based on their putative functions into one of the following categories (Fig. 5): ATP-/GTPases (four), cytoskeletal (seven), cell adhesion (three), kinases and phosphatases (11), RNA processing (10), protein processing (nine), transcription regulation (15), transporters (three), novel/unknown function (13), and other (13). Examples of genes placed in the “other” category included genes involved in cellular respiration, vesicle transport, and signal transduction. Note that every category included in Figure 5 was represented by several “hits.” Table 5 contains a complete list of the insertion loci, categorized by probable gene function.
Figure 5.
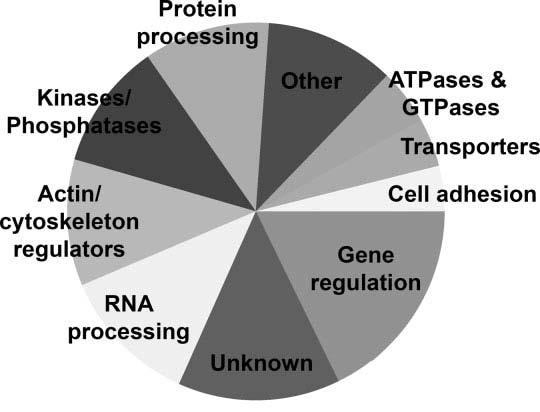
Pie chart showing putative gene functions of insertion loci in mutants identified by the screen. Several different types of proteins are encoded by the insertion loci: four genes are ATP- or GTPases, three may regulate cell adhesion, nine are cytoskeletal molecules, nine are kinases or phosphatases, 10 are involved in RNA processing, nine are involved in protein processing, 15 are transcriptional regulators, three are transporters, nine are involved in other cellular functions, and 12 are novel genes of unknown function.
Table 5.
Genes Affected in Morphology Mutants
| Allele | Phenotype | Affected Gene | Complementation Test | |
|---|---|---|---|---|
| Actin/cytoskeletal regulators: | ||||
| P{GT1}BG00406 | Overgrowth | jbug | Df(2R)59AD | C |
| P{GT1}BG01766 | Undergrowth | dbo | Df(3L)th102 | C |
| P{SUPor-P}mael[KG03309a] | Overgrowth | mael | maelr15 | C |
| P{SUPor-P}nuf[KG02305] | Overgrowth | nuf | P{SUPor-P} nufKG00314 | F |
| P{SUPor-P}KG03611 | Undergrowth | CG33188 | Df(3R)by62 | C |
| P{SUPor-P}crb[KG05098] | PF/TR | crb | Df(3R)Exel6199 | F |
| P{SUPor-P}KG06321 | Overgrowth | ctp | ND | |
| P{SUPor-P}CG2095[KG02723] | Overgrowth | sec8 | Df(3R)Tpl10 | C |
| P{SUPor-P}Mys45A[KG00940] | PF/TR | Mys45A | Df(2R)w45-30n | F |
| ATPases & GTPases: | ||||
| P{GT1}Rgl[BG02025] | Undergrowth | rgl | Df(3L)fz-GF3b | F |
| P{SUPor-P}CG8351[KG01477] | Overgrowth | CG8351 | P{SUPorP}CG8351[KG09501] | F |
| P{SUPor-P}didum[KG04384] | Undergrowth | didum | Df(2R)Drlrv22 | C |
| P{SUPor-P}blw[KG05893] | Overgrowth | blw | P{PZ}blw1 | F |
| Cell adhesion molecules: | ||||
| P{SUPor-P}Ten-m[KG00101] | Undergrowth | Ten-m | Df(3L)Exel6138 | F |
| P{SUPor-P}KG04279 | Overgrowth | ed | Df(2L)Exel8010 | C |
| P{SUPor-P}Sdc[KG06163] | Overgrowth | Sdc | P{PZ}Sdc10608 | C |
| Kinases & phosphatases: | ||||
| P{GT1}Pka-C1[BG02142] | Undergrowth | Pka-C1 | Df(2L)30A-C | C |
| P{SUPor-P}CycE[KG00239] | Overgrowth | CycE | CycEAR95 | F |
| P{SUPor-P}l(3)00305[KG01077] | Undergrowth | l(3)00305 | P{PZl(3)003050030} | F |
| P{SUPor-P}gish[KG03891] | Overgrowth | gish | P{PZ}gish04895 | F |
| P{SUPor-P}PNUTS[KG00572] | Overgrowth | PNUTS | Df(2L)Exel6003 | F |
| P{SUPor-P}JIL-1[KG02848] | Undergrowth | JIL-1 | P{SUPor-P}JIL-1Scim | F |
| P{SUPor-P}polo[KG03033] | Undergrowth | Polo | P{lacW}l(3)S030302 | C |
| P{SUPor-P}IP3K1[KG02192] | Overgrowth | IP3K1 | P{PZ} IP3K1UY530 | C |
| P{SUPor-P}KG06341 | Overgrowth | KP78a/Pros | Df(3R)M-Kx1 | F |
| RNA processing: | ||||
| P{GT1}cpo[BG02810] | Overgrowth | cpo | Df(3R)sr16 | F |
| P{GT1}SmB[BG02775] | PF/TR | SmB | Df(2L)J2 | F |
| P{SUPor-P}Pabp2[KG02359] | PF/TR | Pabp2 | Df(2R)Exel17095 | F |
| P{SUPor-P}KG01427 | Undergrowth | CG8268 | Df(3L)ZP1 | F |
| P{SUPor-P}KG03372 | Overgrowth | CG5933 | Df(3R)crb-F89-4 | C |
| P{SUPor-P}armi[KG04664] | Undergrowth | armi | Df(3L)1227 | C |
| P{SUPor-P}KG07049 | Overgrowth | REF2 | Df(2L)Prl | F |
| P{SUPor-P}KG04415 | PF/TR | tsu | Df(2R)G53 | F |
| P{SUPor-P}KG00080 | Overgrowth | ARC32 | Df(2R)Exel7162 | C |
| P{SUPor-P}CG8545[KG08631] | Overgrowth | CG8545 | Df(2R)BSC3 | F |
| Protein processing: | ||||
| P{GT1}tara[BG01673] | PF/TR | tara | Df(3R)sbd45 | F |
| P{GT1}Rpn11[BG01694] | Overgrowth | Rpn11 | Df(2L)sc19-7 | C |
| P{SUPor-P}KG02920 | Undergrowth | hsromega | ND | |
| P{SUPor-P}KG03291 | Undergrowth | pgant5 | Df(2L)sc19-6 | C |
| P{SUPor-P}CG10882[KG02906] | Overgrowth | CG10882 | EP(2)2058 | F |
| P{SUPor-P}Tim10[KG04105] | Undergrowth | Tim10 | P{SUPorP}Tim10KG04117b | C |
| P{SUPor-P}CG15266[KG01622] | Overgrowth | CG15266 | Df(2L)osp18 | C |
| P{SUPor-P}KG03071 | Overgrowth | Nedd8 | P{EP}EP2063 | F |
| P{SUPor-P}KG05649a | Overgrowth | rt | rt2 | C |
| Gene regulation: | ||||
| P{GT1}simj[BG00403] | Overgrowth | CG32067 | Df(3L)BSC14 | C |
| P{GT1}Trap100[BG01670] | Undergrowth | Trap100 | Df(3L)Exel6112 | F |
| P{GT1}psq[BG02315] | Undergrowth | psq | psqX1-36 | C |
| P{SUPor-P}Ssdp[KG03600] | Overgrowth | Ssdp | SsdpL5 | F |
| P{SUPor-P}CtBP[KG00309] | Overgrowth | CtBP | CtBP03463 | F |
| P{SUPor-P}Kr-h1[KG00354] | Overgrowth | Kr-h1 | ND | |
| P{GT1}ftz-f1[BG01734] | Overgrowth | ftz-f1 | Df(3R)CP1 | C |
| P{GT1}chif[BG02820] | Overgrowth | chif | Df(2L)Exel7065 | C |
| P{SUPor-P}CCR4[KG00877]a | PF/TR | CCR4 | Df(3R)Exel6198 | F |
| P{SUPor-P}gcm[KG01117] | Undergrowth | gcm | Df(2L)Exel7042, | F |
| P{SUPor-P}KG02638 | Overgrowth | Hr46 | P{SUPor-P}Hr46KG00850 | C |
| P{SUPor-P}dimm[KG02598] | Overgrowth | dimm | dimm929 | C |
| P{SUPor-P}KG02164 | PF/TR | scrt | Df(3L)CH39 | C |
| P{SUPor-P}KG06157 | Overgrowth | tna | P{lacW}tnaL6731 | F |
| P{SUPor-P}KG01549 | Undergrowth | jing | P{lacW}jingk03404 | F |
| Transporters: | ||||
| P{SUPor-P}CG10365[KG00107] | PF/TR | CG10365 | Df(3R)Exel9013 | C |
| P{SUPor-P}CG5226[KG03347] | Overgrowth | CG5226 | Df(2R)PC4 | C |
| P{SUPor-P}Scim13[1] | Undergrowth | Scim13 | Df(2L)N6 | C |
| Other: | ||||
| P{GT1}BG00476 | Undergrowth | CG4860 | Df(3R)E79 | F |
| P{GT1}sg1[BG02666] | Undergrowth | sgl | Df(3L)XDI98 | F |
| P{GT1}l(2)35Di[BG02008] | Overgrowth | l(2)35Di | Df(2L)osp29 | C |
| P{SUPor-P}KG01429 | Overgrowth | Mrps6 | Df(3L)Exel8098 | F |
| P{SUPor-P}KG02500 | Overgrowth | Gyc76C | Df(3L)Exel6135 | C |
| P{SUPor-P}CG8552[KG03914] | Overgrowth | CG8552 | In(1)wm4h | C |
| P{SUPor-P Cop[KG06383] | Other | γCop | P{lacW}γCopS057302a | F |
| P{SUPor-P}Scim15[1] | Overgrowth | Scim15 | Df(2L)Mdh, | C |
| P{SUPor-P}Hr39[Scim] | Overgrowth | Hr39 | P{GT1}Hr39BG02396 | C |
| Unknown: | ||||
| P{GT1}BG00531 | PF/TR | unknown | Df(3R)Exel6141 | C |
| P{GT1}BG01610 | Undergrowth | unknown | ND | |
| P{SUPor-P}KG00670 | PF/TR | unknown | ND | |
| P{SUPor-P}KG03146 | Undergrowth | unknown | Df(3R)Exel6220 | F |
| P{SUPor-P}KG03188 | PF/TR | CG13842 | Df(2R)BSC3 | C |
| P{SUPor-P}KG01736 | Overgrowth | CG14363 | Df(3R)red3l | C |
| P{SUPor-P}KG00158 | Overgrowth | CG3793 | ND | |
| P{SUPor-P}l(1)G0003[KG02485] | Overgrowth | CG6606 | Df(1)E128 | C |
| P{SUPor-P}CG7728[KG00300] | PF/TR | CG7728 | Df(3L)Exel7253 | F |
| P{SUPor-P}l(3)61Da[286-4] | Overgrowth | l(3)61Da | Df(3L)Ar14-8 | F |
| P{SUPor-P}oaf[Scim-a] | Overgrowth | oaf | P{SUPor-P}oafKG00814 | C |
| P{SUPor-P}Scim12[2] | Overgrowth | Scim12 | Df(2L)Exel6277 | C |
These results suggest that many different protein types are required for normal NMJ development; no particular type of protein seems to dominate. Therefore, synaptogenesis is likely dependent on multiple aspects of cell function.
Mutant Gene Expression Patterns
The expression patterns of many of the insertion loci identified by our study have been analyzed in previous studies or as part of the BDGP embryonic gene expression project, which has determined embryonic gene expression patterns of approximately one-fifth of the Drosophila genome by in situ hybridization (Tomancak et al., 2002). Because all of the mutants identified in our screen affect motor neuron growth and/or differentiation, we expected that most of the insertion loci would be expressed primarily in the nervous system and/or the somatic musculature. Of the 88 mutants identified in our screen, expression patterns for 42 of the insertion loci have been determined (Table 4). Twenty-two of these genes are expressed in the nervous system. Three of these genes are expressed in muscle; 8 are expressed ubiquitously; 9 are primarily expressed in other tissues, including germ cells, gut, or the visual system. Of the insertion loci with known expression patterns, therefore, 78% are expressed in neurons and/or muscles (as expected). The other 22% of genes may act noncell autonomously, or the NMJ phenotypes we observed may be not be due to the transposon insertion (see below).
Table 4.
Expression Patterns of Insertion Loci
| Nervous System | Muscle | Ubiquitous | Other | Unknown | |
|---|---|---|---|---|---|
| Number of lines | 22 | 3 | 5 | 8 | 45 |
| Percentage (of known) | 58 | 8 | 13 | 21 | - |
Complementation Analysis of Synaptic Morphology Mutants
The P-element mobilizations that generated each of the P-element alleles for the Gene Disruption Project may have created second-site mutations that are not tagged by an insertion. Spontaneous mutations can also easily be stabilized and propagated in a stock carrying a balanced lethal mutant chromosome such as those screened here. Background mutations such as these could contribute to the phenotypes identified in our screen and generate false positives, shown in Table 5. To estimate the frequency of lethal background mutations in this particular collection of mutant stocks, we performed complementation tests to determine whether the P-element insertions failed to complement lethal alleles of the same insertion locus.
Of the 88 mutants identified in our screen, 79 were tested for complementation to either (1) independently generated lethal alleles of the locus carrying the insertion, or (2) deficiencies that remove the insertion site (Table 5). Thirty-eight (49%) of the seventy-seven complementation tests failed, suggesting that the lethality of slightly over half of the mutants was caused by a mutation other than the P-element insertion. This result is not likely due to incorrect annotation of available deficiencies; we repeated all tests that complemented, with different deficiencies (not listed), and achieved identical complementation results.
RT-PCR Validation of a Subset of Morphology Mutants
GT1 and SuPor-P P-element insertions are thought to generate loss-of-function phenotypes by disrupting expression or structure of the insertion loci transcripts. Therefore, each insertion mutant should show loss of transcript or alteration in transcript size, as evaluated by RT-PCR. We tested whether this was true for a subset of mutants, selected from each of the major categories shown in Table 5. Three insertion loci showed very strong or complete loss of transcript, as expected: ed, Rpn11, and CG5226. However, four insertion loci showed no reduction in transcript abundance or difference in transcript size: mael, CG8545, CG8351, and CG4860. This supports the idea that lethality in many Gene Disruption Project P-element mutants is due to a background mutation and not due to the transposon insertion. Interestingly, several of these background lethal mutations appear to have become suppressed over the course of the study (presumably due to accumulation of mutations suppressing lethality); all adult flies in all 88 lines isolated by the screen originally carried balancer markers according to BDGP Gene Disruption Project annotations and independent verification of such by the labs of D. Featherstone and B. McCabe. At present, however, (≈1.5 years later), at least some adult flies in five of 88 lines (6%) are missing balancer markers. Although comprehensive validation has not yet been completed, we have yet to observe apparent suppression of NMJ phenotypes.
Our RT-PCR results also support the idea that NMJ phenotypes in at least some mutants may not be due to the insertion. There was no apparent correlation between loss of transcript and lethality complementation failure, suggesting that lethality and NMJ phenotypes were relatively independent and that there is no way to know which insertions cause NMJ phenotypes without extensive validation for each gene. As a result, we urge caution when interpreting the list of genes in Table 5, and instead stress the larger-scale genomic conclusions that can be safely derived from our experiments.
DISCUSSION
Our goal was to use the growing collection of P-element transposon insertion mutants generated by the Berkeley Drosophila Gene Disruption Project to identify genes required for formation and morphological development of the Drosophila NMJ. The prospect of screening Gene Disruption Project mutants was attractive for several reasons: the mutagenesis is already completed, mutant lines exist as stocks, all maintained lines carry unique gene insertions (thus avoiding the same locus being repeatedly identified), and the insertion site for each mutant has already been identified via inverse PCR. Unfortunately, our complementation tests and a limited set of RT-PCR assays suggest that this approach requires caution. Approximately half of the transposon mutants appear to carry background lethal mutations, and the insertion locus may not be functionally disrupted in some lines. Perhaps this is not surprising; a P-element mutagenesis, like any other mutagenesis, is likely to create multiple genomic alterations even if any particular mutant chromosome ultimately carries a single insertion. The existence of a “tagged” gene clearly does not imply that ordinary genetic precautions can be discarded. We do not wish to discount the value of the growing Gene Disruption Project mutant collection; by definition, all of the BDGDP insertion loci are mutated, and these insertions are excellent tools for generation of specific gene knock-outs via transposon excision, or markers for recombination mapping (Roberts, 1998; Zhai et al., 2003). Candidates listed in Table 5 should be validated by transgene rescue, replication of the phenotype by heteroallelic animals, and/or precise excision with reversion of the phenotype before undue emphasis is placed on any particular gene. Nevertheless, we can draw important conclusions from our data, as follows.
The number of genes encoding proteins required for motor axon outgrowth and pathfinding appears to be small, based on the fact that relatively few of this mutant class were identified. This apparent low incidence suggests one or more of the following possibilities:
The multiple cues required for axon pathfinding and target recognition are encoded by only a few genetic loci, with diversity generated by (for example) alternate splicing and/or posttranscriptional modification, as has been proposed for Dscam (Schmucker and Flanagan, 2004; Wojtowicz et al., 2004; Zhan et al., 2004).
Guidance and target recognition choices are not made using multiple recognition cues, but rather are hierarchical choices—paths and potential targets are chosen via a limited series of increasingly restrictive binary choices. Hierarchy requires many fewer recognition molecules for selection among many paths and targets. Hierarchy is demonstrated by the ability of even-skipped to control whether Drosophila peripheral axons grow to ventral or dorsal regions of the neuromusculature (Landgraf et al., 1999; Labrador et al., 2005).
Finally, it is a possibility that axon guidance and target recognition genes function with so much redundancy that mutation of a single gene is unlikely to cause highly penetrant phenotypes easily recognizable in a screen.
We recovered a surprisingly large number of “overgrowth” mutants in our screen—approximately half of all mutants with a NMJ phenotype showed a qualitative increase in the number of NMJ branches and boutons. This suggests that NMJ growth is actively limited by the action of many different proteins, and therefore normal NMJ morphology is likely a dynamic balance between processes promoting growth, and processes inhibiting growth.
Overall, the number of mutants identified in our screen was surprisingly large; 88 of 202 (44%) mutants examined in detail showed NMJ phenotypes. This suggests that as much as 44% of the genome may be required for normal NMJ formation and development. This assumes, based on our subsequent complementation, RT-PCR, and suppression data, that the initial selection for lethality did little to enrich for NMJ phenotypes. Arguing against this are mutants such as those shown in Figure 2, which lack NMJs and are thus paralyzed and lethal. If we assume that the initial selection for lethality enriched perfectly for mutants with NMJ phenotypes, then our estimate changes to 88/2185 = 4% of the genome. But there are also mutants with dramatic NMJ phenotypes, such as hiw (Wan et al., 2000), which are adult viable. Therefore, the true value likely lies between these two extremes: 4-44% of the genome appears to be required for normal NMJ formation. In any case, this is a surprisingly large number of genes.
Our screen identified many types of genes previously implicated in synapse formation. For example, our screen identified several cell-adhesion molecules. Cell adhesion molecules such as cadherins (Shapiro and Colman, 1999; Bruses, 2000), integrins (Einheber et al., 1996; Anton et al., 1999), and neurexin and neuroligins (Ichtchenko et al., 1996; Missler and Sudhof, 1998; Scheiffele et al., 2000; Chih et al., 2005; Nam and Chen, 2005) are involved in the identification of target cells by the growth cone. Our screen also identified a number of genes encoding cytoskeletal proteins. The cytoskeletal molecule, actin, is localized to the growth cone. Guidance cues bind to receptors to regulate the actin cytoskeleton and control growth cone trajectory via regulatory signaling from Rho GTPases (Gallo and Letourneau, 2004). In the Drosophila NMJ, proteins are anchored to synaptic actin and spectrin cytoskeletons (Featherstone et al., 2001; Chen et al., 2005). One of the largest groups of genes identified by our screen was comprised of kinases and phosphatases. A number of kinases are known to be important during synaptogenesis. During synaptogenesis at the mammalian NMJ, the presynaptic cell releases agrin, which clusters acetylcholine receptors via the muscle specific kinase MuSK (Willmann and Fuhrer, 2002). The EphB2 receptor tyrosine kinase has been shown to both phosphorylate syndecan-2, a cell adhesion molecule (Ethell et al., 2001) and cluster NMDA receptors (Dalva et al., 2000). Most of the rest of the genes that we identified appear to be involved in transcription, translation, and protein processing. Transcription, translation, and protein processing are likely to be involved both upstream and downstream of ATP- and GTPases, cell adhesion proteins, cytoskeletal molecules, and signaling cascades mediated by kinases and phosphatases. Furthermore, our screen identified several genes that were identified in previous Drosophila screens for cell morphology and/or NMJ formation mutants, including CycE, polo, Ten-m (Kiger et al., 2003), and JIL-1 (Kraut et al., 2001). Thus, the genes identified by our screen are consistent with previous studies and what might be expected. Nevertheless, we again urge caution when considering any particular entry in Table 5 without further validation.
We can more reliably consider the types of genes identified in our screen. Each of the protein categories depicted in Figure 5 is represented by several insertion loci. Therefore, even assuming a 50% successful “hit rate” (this is a worst-case scenario; the true success rate is likely to be much higher, based on expression data and ongoing validation), the categories and relative fraction of genes in each category is likely valid. This “survey of the genomic landscape” is analogous to flying over new terrain in a plane, sacrificing detail for a broad perspective that will best enable us to later focus in detail on areas of most interest. This perspective suggests Drosophila NMJ development involves a relatively small number of cell adhesion molecules, extensive networks of transcriptional and translational control, multitudinous intracellular signaling, cytoskeletal rearrangement, and—most importantly—a large fraction of proteins with unknown function. A focus on these areas will likely best lead to important advances in our understanding of synaptogenesis.
In summary, we screened approximately 2185 P-element mutants (representing insertions in 16% of the entire Drosophila genome) for lethal mutants with defects in NMJ development. Embryonic/larval NMJs in 202 lethal mutants (≈1.5% of the genome) were examined in detail using immunocytochemistry and confocal microscopy. We found 88 mutations that affect NMJ morphology. Subsequent control experiments revealed that many of the transposon insertion mutants in the BDGDP carry background mutations, but our data still offer a larger-scale, genome-scale perspective. This view suggests that successful presynaptic growth and differentiation of NMJs appears to require a precisely balanced orchestration of most aspects of cell biology, including many proteins with functions yet to be discovered.
Footnotes
Contract grant sponsor: NIH NINDS; contract grant number: R01 NS045628 (D.F.).
Contract grant sponsor: ALSA (B.D.M.).
REFERENCES
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. Wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Ackley BD, Jin Y. Genetic analysis of synaptic target recognition and assembly. Trends Neurosci. 2004;27:540–547. doi: 10.1016/j.tins.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999;22:277–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. Transposons reanimated in mice. Cell. 2005;122:322–325. doi: 10.1016/j.cell.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Broadie K. Forward and reverse genetic approaches to synaptogenesis. Curr Opin Neurobiol. 1998;8:128–138. doi: 10.1016/s0959-4388(98)80016-0. [DOI] [PubMed] [Google Scholar]
- Bruses JL. Cadherin-mediated adhesion at the interneuronal synapse. Curr Opin Cell Biol. 2000;12:593–597. doi: 10.1016/s0955-0674(00)00137-x. [DOI] [PubMed] [Google Scholar]
- Chen K, Merino C, Sigrist SJ, Featherstone DE. The 4.1 protein coracle mediates subunit-selective anchoring of Drosophila glutamate receptors to the postsynaptic actin cytoskeleton. J Neurosci. 2005;25:6667–6675. doi: 10.1523/JNEUROSCI.1527-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Einheber S, Schnapp LM, Salzer JL, Cappiello ZB, Milner TA. Regional and ultrastructural distribution of the alpha 8 integrin subunit in developing and adult rat brain suggests a role in synaptic function. J Comp Neurol. 1996;370:105–134. doi: 10.1002/(SICI)1096-9861(19960617)370:1<105::AID-CNE10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K. Functional development of the neuromusculature. In: Gilbert L, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science. Vol. 2. Pergamon Press; New York: 2004. pp. 85–134. [Google Scholar]
- Featherstone DE, Davis WS, Dubreuil RR, Broadie K. Drosophila alpha- and beta-spectrin mutations disrupt presynaptic neurotransmitter release. J Neurosci. 2001;21:4215–4224. doi: 10.1523/JNEUROSCI.21-12-04215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Rushton E, Rohrbough J, Liebl F, Karr J, Sheng Q, Rodesch CK, et al. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J Neurosci. 2005;25:3199–3208. doi: 10.1523/JNEUROSCI.4201-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Rushton EM, Hilderbrand-Chae M, Phillips AM, Jackson FR, Broadie K. Presynaptic glutamic acid decarboxylase is required for induction of the postsynaptic receptor field at a glutamatergic synapse. Neuron. 2000;27:71–84. doi: 10.1016/s0896-6273(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58:92–102. doi: 10.1002/neu.10282. [DOI] [PubMed] [Google Scholar]
- Goda Y, Davis GW. Mechanisms of synapse assembly and disassembly. Neuron. 2003;40:243–264. doi: 10.1016/s0896-6273(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Gramates LS, Budnik V. Assembly and maturation of the Drosophila larval neuromuscular junction. Int Rev Neurobiol. 1999;43:93–117. doi: 10.1016/s0074-7742(08)60542-5. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Chiba A, Johansen J, Keshishian H. Growth cone behavior underlying the development of stereotypic synaptic connections in Drosophila embryos. J Neurosci. 1991;11:3227–3238. doi: 10.1523/JNEUROSCI.11-10-03227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch M, Goodman CS. Cell and substrate adhesion molecules in Drosophila. Annu Rev Cell Biol. 1991;7:505–557. doi: 10.1146/annurev.cb.07.110191.002445. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Johansen J, Halpern ME, Johansen KM, Keshishian H. Stereotypic morphology of glutamatergic synapses on identified muscle cells of Drosophila larvae. J Neurosci. 1989;9:710–725. doi: 10.1523/JNEUROSCI.09-02-00710.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian H, Chiba A, Chang TN, Halfon MS, Harkins EW, Jarecki J, Wang L, et al. Cellular mechanisms governing synaptic development in Drosophila melanogaster. J Neurobiol. 1993;24:757–787. doi: 10.1002/neu.480240606. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R, Menon K, Zinn K. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr Biol. 2001;11:417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- Labrador JP, O’Keefe D, Yoshikawa S, McKinnon RD, Thomas JB, Bashaw GJ. The homeobox transcription factor Even-skipped regulates netrin-receptor expression to control dorsal motor-axon projections in Drosophila. Curr Biol. 2005;15:1413–1419. doi: 10.1016/j.cub.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron. 1999;22:43–52. doi: 10.1016/s0896-6273(00)80677-7. [DOI] [PubMed] [Google Scholar]
- Laviolette MJ, Nunes P, Peyre JB, Aigaki T, Stewart BA. A Genetic Screen for Suppressors of Drosophila NSF2 Neuromuscular Junction Overgrowth. Genetics. 2005;170:779–792. doi: 10.1534/genetics.104.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl FLW, Chen K, Karr J, Sheng Q, Featherstone DE. Increased synaptic microtubules and altered synapse development in Drosophila sec8 mutants. BMC Biology. 2005;3:27. doi: 10.1186/1741-7007-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe BD, Hom S, Aberle H, Fetter RD, Marques G, Haerry TE, Wan H, et al. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004;41:891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc Natl Acad Sci USA. 2005;102:6137–6142. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen KW, Nordeen EJ. Synaptic and molecular mechanisms regulating plasticity during early learning. Ann NY Acad Sci. 2004;1016:416–437. doi: 10.1196/annals.1298.018. [DOI] [PubMed] [Google Scholar]
- Parnas D, Haghighi AP, Fetter RD, Kim SW, Goodman CS. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron. 2001;32:415–424. doi: 10.1016/s0896-6273(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Roberts DB, editor. Drosophila: A Practical Approach. 2nd ed. Oxford University Press; Oxford: 1998. [Google Scholar]
- Rose D, Chiba A. A single growth cone is capable of integrating simultaneously presented and functionally distinct molecular cues during target recognition. J Neurosci. 1999;19:4899–4906. doi: 10.1523/JNEUROSCI.19-12-04899.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, Geyer PK. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Apel ED, Burgess RW, Emerson RB, Feng G, Gautam M, Glass D, et al. Development of the neuromuscular junction: genetic analysis in mice. J Physiol Paris. 1998;92:167–172. doi: 10.1016/s0928-4257(98)80004-1. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Flanagan JG. Generation of recognition diversity in the nervous system. Neuron. 2004;44:219–222. doi: 10.1016/j.neuron.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Sink H, Whitington PM. Pathfinding in the central nervous system and periphery by identified embryonic Drosophila motor axons. Development. 1991;112:307–316. doi: 10.1242/dev.112.1.307. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HI, DiAntonio A, Fetter RD, Bergstrom K, Strauss R, Goodman CS. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Willmann R, Fuhrer C. Neuromuscular synaptogenesis: clustering of acetylcholine receptors revisited. Cell Mol Life Sci. 2002;59:1296–1316. doi: 10.1007/s00018-002-8509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Kawano T, Weimer RM, Bessereau JL, Zhen M. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J Neurosci. 2005;25:3833–3841. doi: 10.1523/JNEUROSCI.4978-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara M, Rheuben MB, Kidokoro Y. Transition from growth cone to functional motor nerve terminal in Drosophila embryos. J Neurosci. 1997;17:8408–8426. doi: 10.1523/JNEUROSCI.17-21-08408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, et al. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci USA. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]


