FIG. 4.
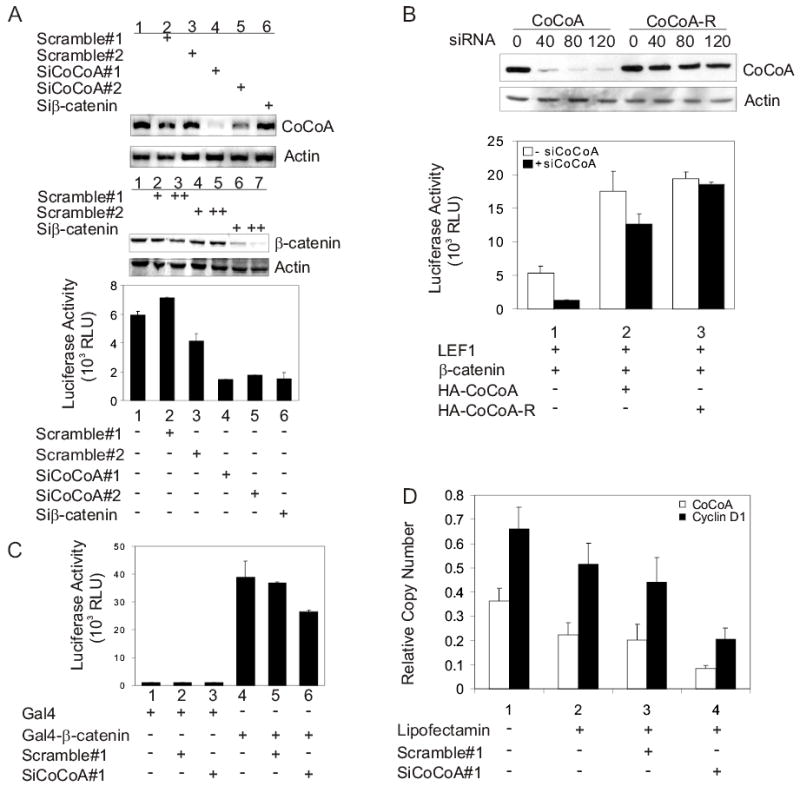
Requirement of endogenous CoCoA for β-catenin and LEF1 function. (A) COS-7 cells were transfected in 12-well plates with pGL3OT reporter plasmid (200 ng), pSG5.HA-LEF1 (100 ng), and either siRNA targeting CoCoA, siRNA with scrambled sequence, or siRNA targeting β-catenin (+=40 pmole, ++=60 pmole). Cell lysates were collected 48 h after transfection and used for luciferase assays (lower panel), for immunoblots detected with anti-β-catenin and anti-actin antibodies (middle panels), or for preparation of total RNA for RT-PCR analysis using primers for CoCoA or β-actin cDNA (upper panels). (B) For immunoblots (upper panels), COS-7 cells were transfected in 12-well plates with 1 μg of either pSG5.HA-CoCoA or pSG5.HA-CoCoA-R and the indicated amount (pmole) of siRNA targeting CoCoA. Expression of HA-CoCoA was analyzed with anti-HA antibodies, and endogenous β-actin was detected with anti-actin antibodies. For transient reporter plasmid assays, CV-1 cells were transfected in 12-well plates with or without siRNA targeting CoCoA (40 pmole) using Lipofectamine 2000. 48 h after siRNA transfection, cells were transfected using Targefect F1 with pGL3OT (200 ng), pSG5.HA-LEF1 (1 ng), pSG5.HA-β-catenin (10 ng), and either pSG5.HA-CoCoA or pSG5HA-CoCoA-R (200 ng) as indicated. Cell lysates were prepared 24 h after plasmid transfection for measurement of luciferase activity. Results shown are representative of two independent experiments. (C) COS-7 cells were transfected in 12-well plates with GK1-LUC reporter plasmid (200 ng), plasmids encoding Gal4 DBD or Gal4DBD-β-catenin (150 ng), and 20 pmole of either siRNA targeting CoCoA or siRNA with scrambled sequence as indicated. Luciferase activity was measured 48 h after transfection. Results shown are representative of four independent experiments. (D) SW480 cells were transfected with 60 pmole of either siRNA targeting CoCoA or siRNA with scrambled sequence as indicated. Total RNA was collected and cDNA was produced for quantitative real-time PCR analysis. Results shown are normalized to GAPDH mRNA levels and are the mean and standard deviation from 4 QPCR reactions for a single transfection experiment, which is representative of six independent experiments. For the six independent experiments, comparing cells treated with siCoCoA versus scrambled sequence siRNA, p=0.004 for CoCoA mRNA and p=0.01 for Cyclin D1 mRNA. Supplementary Fig. S1 shows the results and statistical analysis of all six experiments. In addition Supplementary Fig. S1 shows that the Correlation Coefficient R2 is 0.91 for the 6 experiments when the levels of CoCoA mRNA and Cyclin D1 mRNA are compared in the cells which were treated with siRNA against CoCoA; this indicates that the siRNA against CoCoA causes a proportional decrease in the levels of CoCoA mRNA and Cyclin D1 mRNA.
