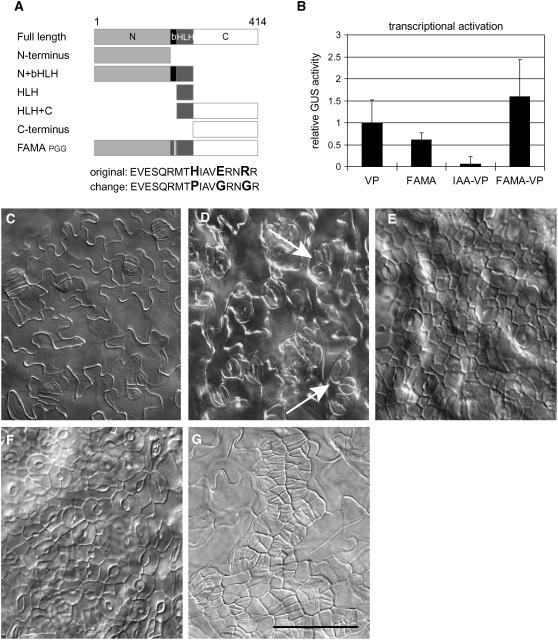
Figure 5.
Behavior of FAMA Domains in Transcriptional Activity Assay and in Planta.
(A) Diagram of deletion and DNA binding site mutation constructs tested.
(B) Protoplast-based transcriptional activation assay. Effector constructs indicated on x axis. All values were normalized to VP16-driven reporter activation values. Error bars indicate standard deviations. n = 6 independent transformations.
(C) to (G) DIC images of cotyledons of plants transformed with PROEST:FAMA protein variants. All images are at the same magnification.
(C) Col plants expressing PROEST:FAMA-EAR.
(D) Col plants expressing PROEST:FAMA C terminus alone. The arrow indicates paired stomata.
(E) and (F) Col plants expressing PROEST:FAMAPGG at 4 and 10 d, respectively.
(G) PROEST:FAMAPGG in fama-1. Bar = 50 μm.