Figure 7.
Construction of a Homologous Expression System for Characterizing the RPS2–RIN4–NDR1 Interaction.
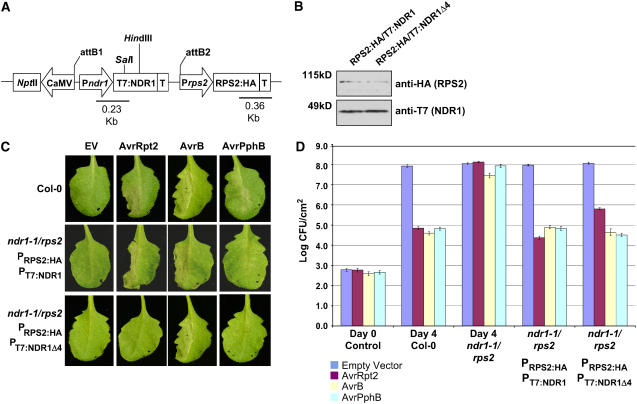
(A) rps2/ndr1-1 mutant Arabidopsis plants were transformed with a dual promoter construct expressing RPS2:HA and T7:NDR1/T7:NDR1Δ4 expressed under the control of their respective native promoters.
(B) Protein gel blot analysis of RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 transgenic plants. Total protein extracts were separated by SDS-PAGE and probed with anti-HA antibodies (RPS2) and anti-T7 (NDR1) to determine protein expression levels in complemented Arabidopsis lines.
(C) High-density (105 colony-forming units [cfu]/mL) inoculation of wild-type Col-0 (top panels), RPS2:HA/T7:NDR1 (middle panels), and RPS2:HA/T7:NDR1 (bottom panels) with P. syringae DC3000 expressing empty vector (EV; control), AvrRpt2, AvrB, and AvrPphB effector proteins.
(D) Bacterial growth curve analysis of native promoter expression lines of Arabidopsis RPS2:HA/T7:NDR1- and RPS2:HA/T7:NDR1Δ4-complemented plants. Bacterial counts were determined at days 0 and 4 following inoculation with P. syringae strains (104 cfu/mL) expressing the indicated effector protein. Day 0 controls were plated 1 h after inoculation and represent the average bacterial growth count of all plant genotypes tested. The Arabidopsis ndr1-1/rps2 genotype corresponds to the parent genotype of the complemented expression system shown in (A). Data shown are the mean of four independent lines for both RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 constructs. Error bars indicate standard deviation calculated from the mean of replicate samples.