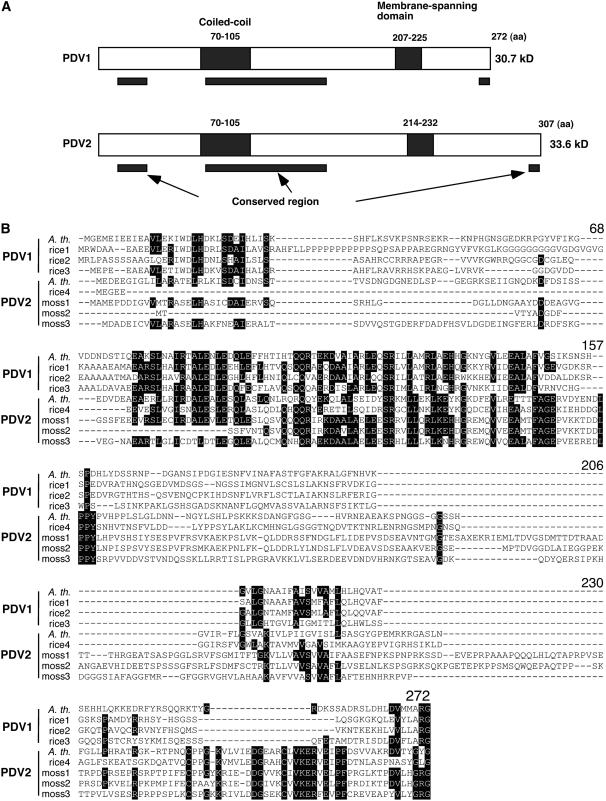
Figure 2.
Predicted Structures of PDV1 and PDV2 Proteins, and Sequence Alignment of PDV1, PDV2, and Homologous Proteins.
(A) Predicted structures of PDV1 and PDV2 proteins. Putative coiled-coil domains and transmembrane domains are depicted as closed rectangles, and their positions within the amino acid sequences are indicated. Black bars below the diagrams delineate the regions that are relatively well conserved among PDV1, PDV2, and related proteins based on the sequence alignment. aa, amino acids.
(B) Sequence alignment of PDV1, PDV2, and related proteins. Black shading indicates residues identical in five or more sequences. The numbers of amino acids from the N terminus of Arabidopsis PDV1 are indicated above the alignment. A. th., Arabidopsis thaliana; rice, Oryza sativa; moss, Physcomitrella patens. Accession numbers for each sequence are listed in Methods. The sequences were aligned with ClustalW (Thompson et al., 1994).