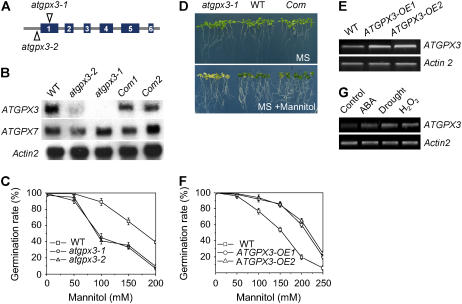
Figure 1.
Characterization of atgpx3 T-DNA Insertion Mutants and Overexpression Transgenic Plants.
(A) The insertion positions of T-DNA in the ATGPX3 gene.
(B) RNA gel blots show ATGPX3 expression in the wild type, atgpx3 mutants, and complemented ATGPX3 (Com) lines. Total RNA was extracted from the wild type, atgpx3-1, atgpx3-2, and complementation lines. Fifteen micrograms of total RNA was loaded in each lane. An Actin gene and ATGPX7 were used as a loading control and a positive control, respectively.
(C) Germination sensitivity to osmotic stress in atgpx3 mutants. Seeds from the wild type, atgpx3-1, and atgpx3-2 were germinated on MS agar medium or MS medium supplemented with different concentrations of mannitol for 7 d. Values are means ± sd of three independent experiments (>120 seeds per point).
(D) Complementation of the phenotype conferred by atgpx3-1 by expression of wild-type ATGPX3. After germinating for 5 d in MS medium, the seedlings were transferred to MS agar medium or MS medium containing 200 mM mannitol and grown for 15 d.
(E) Expression of ATGPX3 in two ATGPX3 overexpression transgenic lines.
(F) Germination insensitivity to osmotic stress in ATGPX3 overexpression transgenic plants. Seeds from the wild type and both overexpression lines were germinated on MS agar medium or MS medium supplemented with different concentrations of mannitol for 7 d. Values are means ± sd of three independent experiments (>120 seeds per point).
(G) RT-PCR analysis of ATGPX3 gene transcripts in response to stress conditions. Arabidopsis seedlings were grown on MS medium plates for 15 d. Wild-type plants were treated with 60 μM ABA, drought stress, or 5 mM H2O2 for 4 h. Total RNA was isolated from treated and untreated wild-type plants. Actin2 primer was used as an internal control.