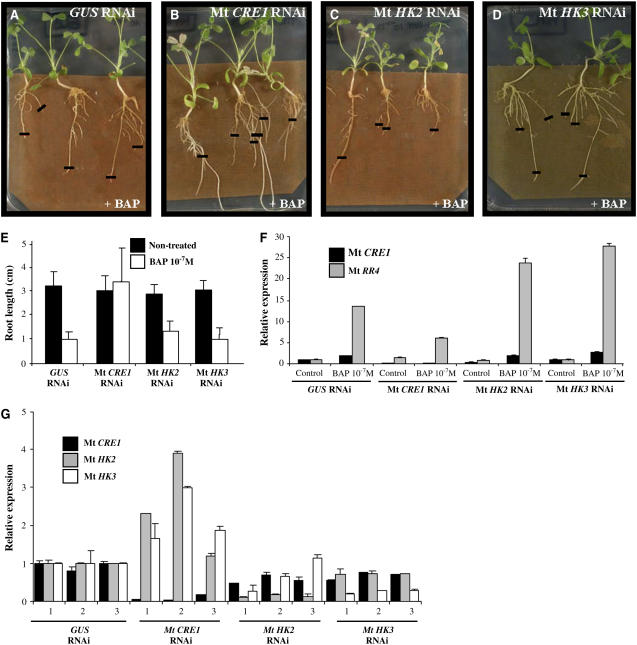
Figure 2.
Mt CRE1 RNAi Roots Are Cytokinin Insensitive.
(A) to (D) Representative examples of root growth observed in A. rhizogenes–transformed M. truncatula roots expressing GUS RNAi, Mt CRE1 RNAi, Mt HK2 RNAi, or Mt HK3 RNAi constructs 6 d after transfer to 10−7 M BAP. Black lines indicate the position of root tips at the moment of transfer. Cytokinin insensitivity is visualized by the root's ability to grow on this inhibitory concentration of cytokinin. The transgenic roots were selected on kanamycin as described by Boisson-Dernier et al. (2001).
(E) Mean length of independent transgenic roots transformed with these different constructs and grown 6 d on media with or without 10−7 M BAP. A representative example out of seven biological experiments is shown (n > 30 per construct and condition), and error bars represent standard deviation (ANOVA, P < 0.001)
(F) Expression analysis of Mt CRE1 and a cytokinin-inducible A-type response regulator, Mt RR4. Real-time RT-PCR analysis was performed on A. rhizogenes–transformed M. truncatula roots carrying either a control (GUS RNAi) or Mt HKs RNAi constructs (n > 15) after a short-term cytokinin treatment (10−7 M BAP for 1 h). Histogram represents the quantification of specific PCR amplification products normalized to the constitutive control Mt ACTIN11. The value of control transgenic roots (i.e., non-BAP-treated GUS RNAi root) is set at 1. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates.
(G) Expression analysis of Mt CRE1, Mt HK2, and Mt HK3. Real-time RT-PCR analysis was performed on independent A. rhizogenes–transformed M. truncatula roots carrying either a control (GUS RNAi) or Mt HK RNAi constructs and grown 7 d on 10−7 M BAP. Histograms represent the quantification of each specific PCR amplification product normalized to the constitutive control Mt ACTIN11. The value of control transgenic roots (i.e., GUS RNAi) is set up at 1 as reference. A representative example out of three biological experiments is shown, and error bars represent standard deviation for three technical replicates.