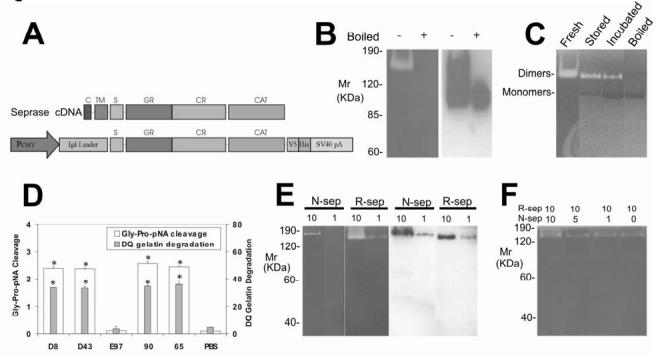
Figure 1Isolation and characterization of r-seprase. A, R-seprase expression vector pE15. The cDNA regions that encode predicted domains of seprase are indicated: C, cytoplasmic domain, amino acids 1-6; TM, transmembrane domain, amino acids 7-26; S, stalk region, amino acids 27-48; GR, glycosylation rich region, amino acids 49-314; CR, cysteine rich domain, amino acids 305-466; CAT, catalytic domain, amino acids 500-760. The cDNA fragment that encodes r-seprase lacks the cytoplasmic and transmembrane domains and was inserted into a CMV promoter driven expression cassette containing an N-terminal mouse Igk secretion signal and a C-terminal V5-His tag. B, A gelatin zymogram (left panel) and a corresponding Western immunoblot (right panel) of r-seprase. The culture medium conditioned by the pE15 transfected 293-EBNA cells was prepared under non-boiled (-) or boiled (+) conditions, and subsequently resolved on a gelatin zymogram and a Western immunoblot using anti-V5 antibody. The purified protein, when boiled for 3 min, completely dissociates into 90-kDa inactive monomers, whereas the proteins remain mostly in dimeric form if not subjected to boiling. The serine gelatinase activity of r-seprase was detected by incubating the zymogram in the presence of 5 mM EDTA to quench metalloprotease activity. Note that both r-seprase dimers and monomers were present in the medium conditioned by transfectants; however, the monomers were gelatinolytically inactive. C, Dissociation of the active 160-kDa r-seprase into the inactive 90-kDa monomer detected by gelatin zymography. Freshly purified r-seprase is active at 160-kDa (Fresh). Moderate activity is detected in the dimerized portion of r-seprase samples stored at -20 °C because a portion has dissociated into 90-kDa inactive monomers (Stored). Samples stored at -20 °C and then subsequently incubated at 37 °C overnight in the presence of 5 mM EDTA also have moderate activity in the portion that remained dimeric; however, a large portion dissociated into 90-kDa inactive monomers (Incubated). Note that r-seprase dimers create white bands against the dark grey background due to their gelatinase activity, whereas the monomers have no gelatinase activity and therefore appear as black bands compared to the gelatin background. D, The proteolytic activities of r-seprase. The seprase specific DP and gelatinase activities were measured using immunocaptured protein in the soluble enzymatic assays under non-denaturing conditions containing 5 mM EDTA as described (15). Seprase was captured from culture medium conditioned with pE15 transfected 293-EBNA cells using rat mAbs D8 and D43 (against n-seprase), mAb E97 (against denatured seprase monomeric subunits and their shortened forms) and mouse mAbs 90 and 65 (against r-seprase dimer). The DP (Gly-Pro-pNA cleavage) and gelatinase (DQ gelatin degradation) activities of the isolated r-seprase were measured in parallel. The symbol * above the bars indicate a significant increase in activity compared with the PBS control (P<0.05). E, Specificity of gelatinase activity of n-seprase and r-seprase revealed by parallel gelatin zymography and Western immunoblotting analyses. 10 and 1 μl of LOX cell Triton X-100 detergent lysate (n-sep) and the concentrated culture medium conditioned by the pE15 transfected 293-EBNA cells (r-sep) were loaded into parallel SDS gels: one was subjected to gelatin zymography in 5 mM EDTA conditions (left panel) for serine-type gelatinase detection and the other by Western immunoblotting using mAb 90 (right panel) for assessment of n- and r-seprase protein. F, Mixing of the r-seprase with different amounts of the detergent extract containing n-seprase to test for the presence of endogenous seprase inhibitors. From 1-10 μl of LOX cell Triton X-100 detergent lysate (n-sep) were mixed with 10 μl of the concentrated culture medium conditioned by the pE15 transfected 293-EBNA cells (r-sep). Mixtures were subjected to gelatin zymography in 5 mM EDTA conditions for assessment of possible inhibition of gelatinase activity specific for r-seprase.
