Figure 4.
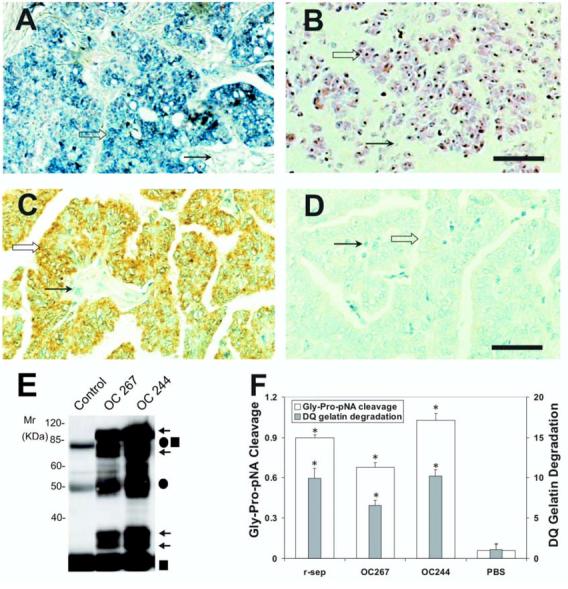
Expression and activity of n-seprase in human ovarian carcinoma. A and B: N-seprase mRNA expression in human ovarian carcinoma tissues. In situ hybridization was performed on formalin-fixed, paraffin-embedded tumor sections using FITC-labeled anti-sense (A) or sense (B) RNA probes for seprase. FITC-labeling was revealed using the alkaline phosphatase colorimetric substrates BCIP/NBT, which show a purple/blue staining. Counterstaining using nuclear fast red appears red. Seprase mRNA is mainly localized in tumor cells (open arrows) and is less prominent in stromal cells (arrows). Bar, 100 μm. C and D: N-seprase protein expression in human ovarian carcinoma tissues. Immunohistochemistry was carried out on formalin-fixed, paraffin-embedded tumor sections using mAb D8 against seprase (C) or a control rat antibody (D). The positive staining is brown, while the counterstaining using hematoxylin appears blue. Seprase protein was mainly detected in tumor cells (open arrows) instead of stromal cells (arrows). Bar, 100 μm. E: Isolation of n-seprase truncated forms from human ovarian carcinoma tissues. Tumor tissues were lysed in the presence of 5mM EDTA. N-seprase in tumor lysates (OC 267 and OC244) was isolated and identified as described in (3B) above. Note that the top two arrows indicate dissociated seprase polypeptides at 97- and 65-kDa; the lower two arrows show dissociated seprase polypeptides at 35- and 25-kDa. F: Seprase specific DP and gelatinase activity in human ovarian carcinoma tissues. Tumor tissue lysates were applied to plates coated with mAb D8 to specifically capture active seprase forms. The soluble enzymatic assays were conducted in the presence of 5 mM EDTA to inhibit any metalloprotease activity. * denotes a significance of P<0.001 compared to the PBS negative control.
