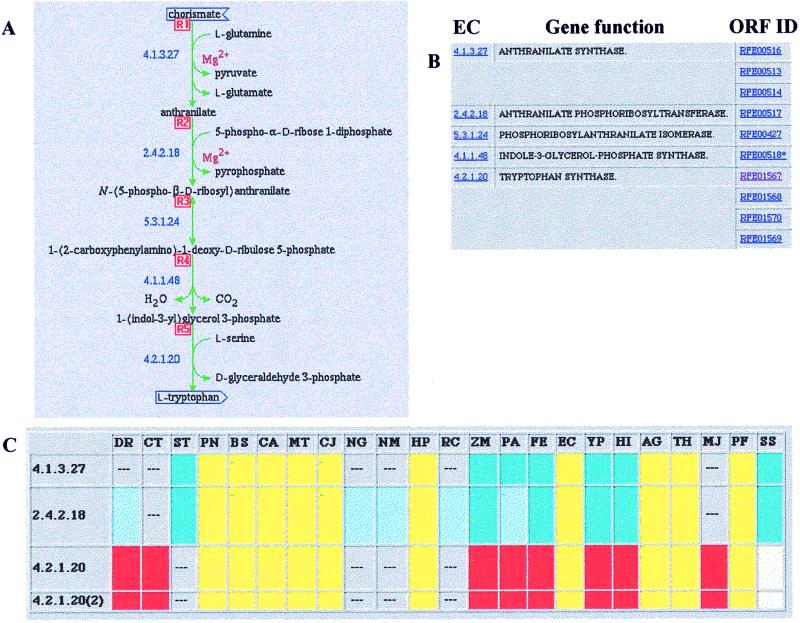
Figure 2.
Tryptophan synthesis in T. ferrooxidans. (A) The substrates and products of different enzymatic reactions together with the EC numbers for the enzymes involved. (B) The EC enzyme numbers, names of the enzymes, and “RFE” ORF assignment number for the gene product in the Thiobacillus database. Any item in blue can be selected on the IG Internet site to reveal deeper information. (C) Functional coupling of genes for tryptophan biosynthesis in different microorganisms. Identical colors indicate genes in corresponding clusters in each organism; the colors are arbitrary. The dashes indicate corresponding assignments not clustered with others in that organism. Note also that the diagram of A and ORF assignments from B can be brought up by way of Fig. 1. DR, Dienococcus radiodurans; CT, cyanobacterium Synechocystis sp.; ST, Streptococcus pyogenes; PN, Streptococcus pneumoniae; BS, Bacillus subtilis; CA, Clostridium acetobutylicum; MT, Mycobacterium tuberculosis; CJ, Campylobacter jejuni; NG, Neisseria gonorrhoeae; NM, Neisseria meningitidis; HP, Helicobacter pylori; RC, Rhodobacter capsulatus; ZM, Zymomonas mobilis; PA, Pseudomonas aeruginosa; FE, Thiobacillus ferrooxidans; EC, Escherichia coli; YP, Yersinia pestis; HI, Haemophilus influenzae; AG, Archaeoglobus fulgidus; TH, Methanobacterium thermoautotrophicum; MJ, Methanococcus jannaschii; PF, Pyrococcus furiosus; and SS, Sulfolobus solfataricus.