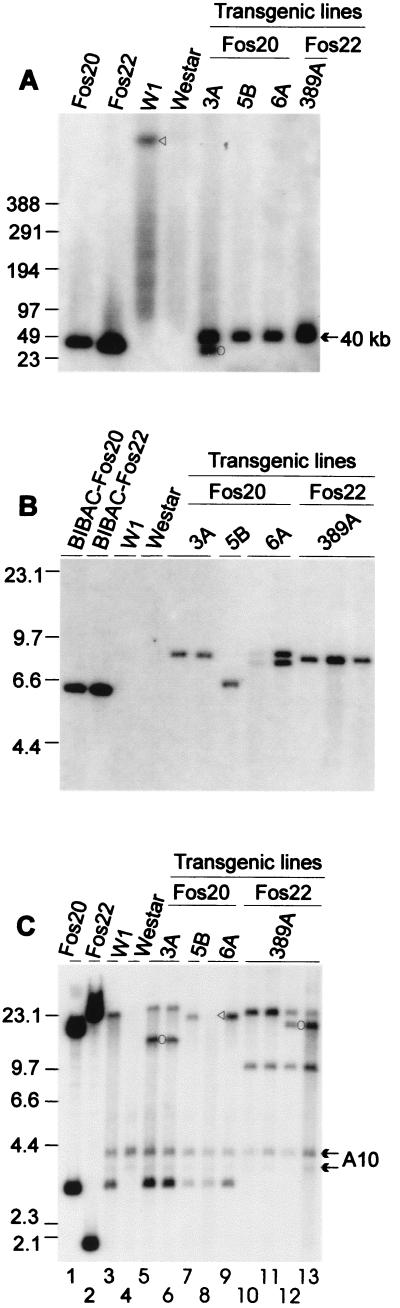
Figure 3.
Genomic DNA gel blot analysis of the T1 progeny plants. Molecular size markers are indicated at left in kilobases. Sources of DNA are labeled above each lane. Each lane represents DNA from one progeny plant. The transgene constructs, W1, and Westar DNAs were all included as controls. (A) Gel-blot hybridization of NotI-digested high molecular mass DNA with SLG-910 and SRK-910 cDNAs. Restriction fragments were separated by pulsed field gel electrophoresis in a CHEF-DRIII apparatus (Bio-RAD) in 1× Tris-acetate-EDTA at 3 V/cm with a pulse-switching interval ramped from 5 to 120 s over 42 h. The triangle indicates the ≈1,000-kb NotI fragment encompassing both the SLG-910 and SRK-910 genes, which was identified previously by pulsed field gel electrophoresis analysis (29). The circle indicates a truncated copy in 3A line. (B) Gel-blot hybridization of HindIII-digested DNA with NPTII-specific DNA. (C) Gel-blot hybridization of EcoRI-digested DNA with SLG-910 and SRK-910 cDNAs. The two fosmids were cut with NotI and EcoRI. The ≈12-kb band in lanes 5 and 6 and the ≈16-kb band in lanes 12 and 13 (indicated by circles) represent truncated copies of the transgenes. The triangle in lane 8 indicates a band visible after longer exposure. The arrows indicate the bands likely representing the endogenous A10 haplotype present in Westar.