Figure 1.
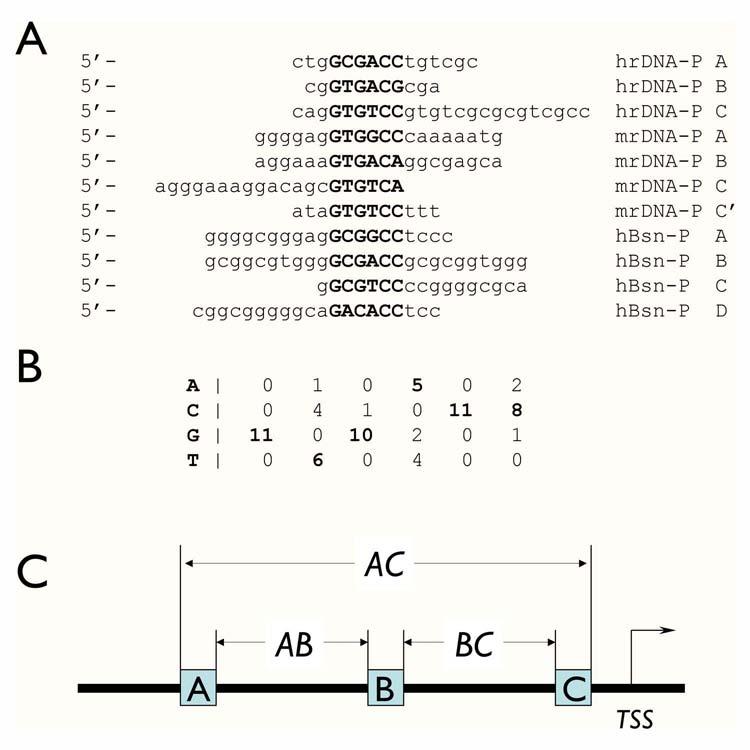
Modeling basonuclin-binding site. A) The eleven Pol I and Pol II promoter sequences, which were protected from DNase I digestion by the N-terminal zinc fingers of basonuclin, were analyzed with software program Consensus, which identifies the statistically most significant sequence matrix, which likely to be the recognition sequence of a protein. The sources of the sequence were: hrDNA-P, human rDNA promoter; mrDNA-P, mouse rDNA promoter; hBsn-P, human basonuclin gene promoter. B) The nucleotide frequency matrix of the basonuclin zinc finger-binding site derived by Consensus. The parameters associated with this matrix were: number of sequences = 11; unadjusted information = 4.79954; sample size adjusted information = 3.8627; ln(p-value) = -33.743; p-value = 2.2162E-15; ln(expected frequency) = -5.68861 expected frequency = 0.00338429. C) The basonuclin binding site module with three basonuclin binding sites constrained by their orientation and spacings (AB and BC). The other defining parameter is the distance of the binding module to the transcription start site (TSS).
