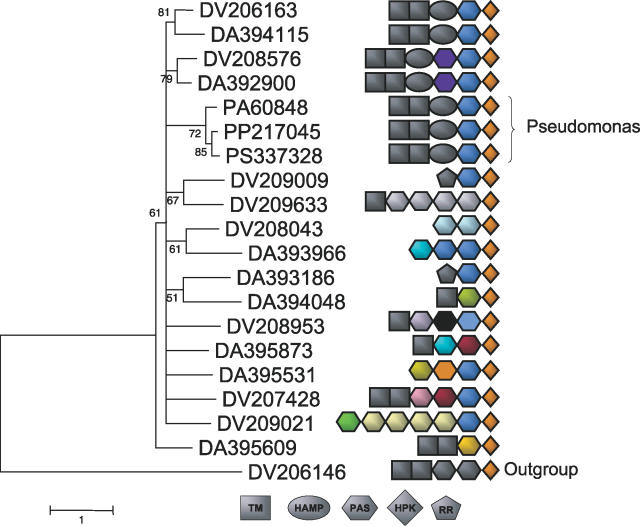
Figure 8. Domain Shuffling in a Desulfovibrio spp. Expansion.
The domain structure of genes in a large LSE in the Desulfovibrio genus is shown. In addition, three similar proteins identified in Pseudomonas species are shown, which are likely the result of HGT. Genes are identified by their species name and their accession number in the MicrobesOnline database (http://microbesonline.org) for easy reference (DV refers to genes present in D. vulgaris and DA refers to genes present in D. alaskensis G20). Each domain corresponds to a branch of the TREE-PUZZLE phylogenetic tree (of only the HPK domains) shown at left. Each PAS domain is colored according to sequence homology (as inferred by BLASTp), and domains with the same color comprise subfamilies of closely related domains. While upstream domains are generally shuffled, each gene shown contains a PAS domain immediately preceding the conserved HPK domain. Moreover, this PAS domain is largely conserved among paralogs at the sequence level, while more N-terminal domains are not. Interestingly, the Pseudomonas gene, which we infer to be involved in a horizontal transfer event, has a set of signaling domains identical to one of the Desulfovibrio copies, suggesting a likely donor–acceptor pair, and highlighting the qualitative difference in genes acquired by HGT and LSE.