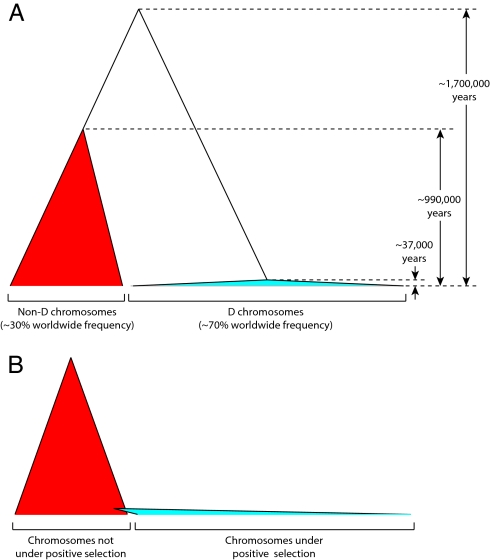
Fig. 2.
Comparison of the microcephalin genealogy with an idealized genealogy. Each filled triangle represents a genealogical clade, with the width of the triangle representing frequency in the population. (A) The genealogy consistent with the haplotype data at the microcephalin locus. The coalescence age of D chromosomes (≈37,000 years), non-D chromosomes (≈990,000 years), and between D and non-D chromosomes (≈1,700,000 years) are indicated. (B) The idealized genealogy of a partial positive selective sweep, wherein the adaptive allele first emerged by a mutational event on a random chromosome in the population.