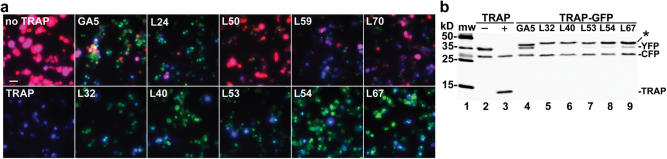
Figure 6.
Screen for linkers that support RNA-binding activity of TRAP. (a) Visual analysis of DsRed2 (pseudocolored red), GFP (pseudocolored green) and CFP (pseudocolored blue) fluorescence in a single field of cells, following fluorescence microscopy and overlay of the three distinct signals. The 293T cells were transfected in 24-well plates with a modified pYIC/5Y-YIC bicistronic reporter gene alone in which the upstream YFP-encoding cistron was replaced with DsRed2 encoding sequences (upper left-hand panel) or co-transfected with expression DNA for TRAP (lower left-hand panel), TRAP–GFP fused through a GA5 linker (upper second panel from left) or TRAP–GFP fused through two different series of linkers with randomized sequences (panels designated with L and a number). Potential positive linkers are those with decreased DsRed2 fluorescence compared to GA5 panel as seen for the bottom panels starting with the second panel on the left. (b) Western blot analysis of potential positive linkers for TRAP–GFP. Ability of TRAP–GFP to selectively repress cap-dependent translation was analyzed using the YFP- and TBS-containing bicistronic reporter gene (pTBS/5Y-YIC). Note the relative level YFP to CFP is lower for the five potential linkers (lanes 5–9) from the visual screen [lower panels of (a)] than the GA5 linker (lane 4). Longer exposure shows that linker L40 (lane 6) appears to fully repress YFP translation, analogous to the level seen for TRAP (lane 3) (data not shown). Asterisk denotes location of TRAP–GFP.