Figure 3.
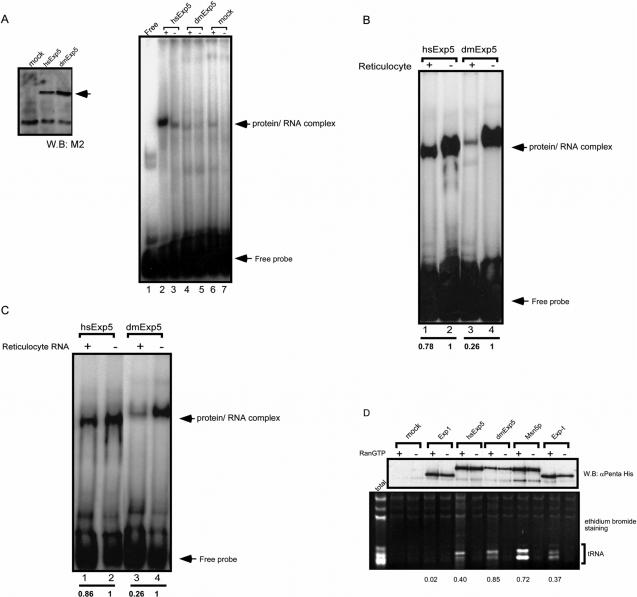
tRNA is a potent inhibitor of the interaction between pre-miR-30 and dmExp5. (A) In vitro translated hsExp5, but not dmExp5, bound pre-miR-30. Left panel: Expression of FLAG-tagged hsExp5 (lane 2) and dmExp5 (lane 3) using an in vitro translation system was confirmed by western blot using anti-FLAG antibody. In lane 1 (mock), an in vitro translation reaction programmed with an empty vector was loaded. Right panel: In vitro translated hsExp5 or dmExp5 as indicated in the left panel was mixed with radio-labeled pre-miR-30 in the presence (+, lanes 2, 4 and 6) or absence (−, lanes 3, 5 and 7) of hsRanQ69L-GTP (lanes 2 and 6) or dmRanQ69L-GTP (lane 4). The RNA–protein complex was detected by EMSA. In lane 1, radio-labeled pre-miR-30 alone was loaded. Arrows indicate the positions of bound and free probe. (B) Bacterially expressed recombinant dmExp5 failed to bind pre-miR-30 in the presence of a reticulocyte lysate. Bacterially expressed recombinant hsExp5 (lanes 1 and 2) or dmExp5 (lanes 3 and 4) along with RanQ69L-GTP were mixed with radio-labeled pre-miR-30 in the presence (indicated by +, lanes 1 and 3) or absence (indicated by −, lanes 2 and 4) of a reticulocyte lysate. The RNA-protein complexes were detected by EMSA. Arrows indicate the positions of bound and free probe. A quantification of the shifted bands was done by using a phosphor imager. The values obtained without the competitor were arbitrary set at 1 and those obtained with the competitor were calculated. The data are indicated below each lane. (C) The binding of dmExp5 to pre-miR-30 was inhibited by RNAs extracted from reticulocyte lysate. Bacterially expressed recombinant hsExp5 (lanes 1 and 2) or dmExp5 (lanes 3 and 4) along with RanQ69L-GTP were mixed with radio-labeled pre-miR-30 in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of an RNA fraction isolated from reticulocyte lysate. The RNA–protein complexes were detected by EMSA. Arrows indicate the positions of bound and free probe. A quantification of the shifted bands was done as in (B). The data are indicated below each lane. (D) Co-immunoprecipitation of tRNAs with exportins. Bacterially expressed recombinant exportins indicated in each lane were mixed with a rabbit reticulocyte lysate in the presence (+) or absence (−) of RanQ69L-GTP or its orthologous mutants from cognate species. After immunoprecipitation with anti-pentaHis antibody-bound Protein G Sepharose, co-precipitated RNAs were extracted from the immunepellets and analyzed by denaturing PAGE followed by ethidium bromide staining (lower panel). Background binding was also examined in the absence of exportins (mock). Co-precipitated proteins were also detected by western blot using an anti-pentaHis antibody (upper panel). Ten percent of input was loaded on the left most lane (total). The amounts of the precipitated exportins and tRNAs in the presence of Ran-GTP were determined by densitometric scanning of the blot and the gel. The relative amounts of co-precipitated tRNAs, which are normalized to the amounts of each exportin, are indicated below the panel.