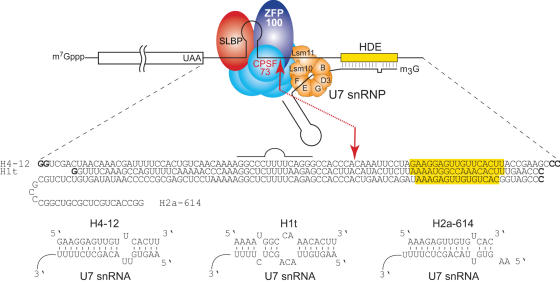
Figure 1.
Schematic representation of the 3′ end processing of replication-dependent histone pre-mRNAs. Cleavage of the RNA occurs at the red arrow between a highly conserved stem–loop structure and the HDE. Trans-acting factors are SLBP, which binds to the hairpin structure, U7 snRNP which anchors the pre-mRNAs by annealing to HDE and the cleavage complex including CPSF-73 which is believed to be the cleavage factor. U7 snRNP contains two U7-specific proteins named Lsm10 and Lsm11. ZFP100 bridges U7 to histone pre-mRNAs by interacting with SLBP and Lsm11. The sequences of the 3′-UTRs used in this work are indicated below the cartoon. Nucleotides shown in bold were added to optimize transcription reactions. Base-pairing of H4-12, H1t and H2a-614 with mouse U7 snRNA is shown at the bottom of the figure.