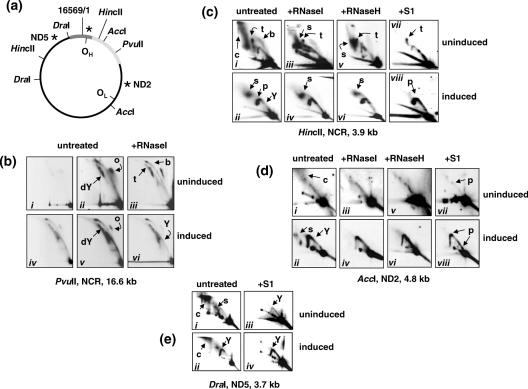
Figure 2.
2DNAGE analysis of mtDNA replication intermediates (RIs) in cells induced to overexpress TFAM-stop. (a) Diagrammatic map of human mtDNA, showing the origins of heavy- (OH) and light-strand (OL) replication according to the orthodox model, relevant restriction sites and probes for the three regions of the genome analysed (approximate location of probes indicated by asterisks). NCR shown as dark grey bar, rDNA as pale grey bar. (b–e) 2DNAGE of mtDNA from uninduced cells and from cells induced to express TFAM-stop for 48 h, analysed using the restriction digests and probes indicated, with or without additional enzymatic treatments as shown. In each panel, the various arcs and other salient features are denoted as follows: Y, standard Y arcs, dY, standard doubleY arcs, c, ‘cloud’ of RNase-sensitive material, o, circular molecules, b, standard bubble arcs, s, slow-moving Y-like arcs, sensitive to various nucleases, t, termination intermediates lying on a portion of a standard X arc, p, prominent pause sites. See Supplementary Figure 5 for diagrammatic interpretations of the various arcs. Panels i and iv of part (b) and panels iii and iv of part (c) are equivalent exposures, for comparison. Other panels of uninduced cell mtDNA are 5- to 10-fold more exposed than induced cell material, in order to reveal the main features of the arcs. Note the general enhancement of RIs, relative loss of nuclease-sensitive species, of termination intermediates and of bubble arcs, following TFAM induction. The appearance of a complete or almost complete, Y arc in Figure 2b, panels iv–vi, is consistent with frequent strand breakage at OH or with recombinational strand-switching (also generating a free end at OH) or with frequent initiation far distant from the NCR.