Abstract
Mobile LTR-retroelements comprising retroviruses and LTR-retrotransposons form a large part of eukaryotic genomes. Their mode of replication and abundance favour the notion that they are major actors in eukaryote evolution. The Gypsy retroelement can spread in the germ line of the fruit fly Drosophila melanogaster via both env-independent and env-dependent processes. Thus, Gypsy is both an active retrotransposon and an infectious retrovirus resembling the gammaretrovirus MuLV. However, unlike gammaretroviruses, the Gypsy Gag structural precursor is not processed into Matrix, Capsid and Nucleocapsid (NC) proteins. In contrast, it has features in common with Gag of the ancient yeast TY1 retroelement. These characteristics of Gypsy make it a very interesting model to study replication of a retroelement at the frontier between ancient retrotransposons and retroviruses. We investigated Gypsy replication using an in vitro model system and transfection of insect cells. Results show that an unstructured domain of Gypsy Gag has all the properties of a retroviral NC. This NC-like peptide forms ribonucleoparticle-like complexes upon binding Gypsy RNA and directs the annealing of primer tRNALys,2 to two distinct primer binding sites (PBS) at the genome 5′ and 3′ ends. Only the 5′ PBS is indispensable for cDNA synthesis in vitro and in Drosophila cells.
INTRODUCTION
Retrotransposons and retroviruses belong to a large family of mobile genetic elements called long terminal repeat (LTR) containing retroelements (1). Ancient retrotransposons, such as yeast transposons (TY) share common genetic features with simple retroviruses, such as vertebrate gammaretroviruses and utilize similar basic mechanisms for their replication. Genome replication proceeds via the conversion of the single-stranded genomic RNA into a double-stranded DNA copy with two LTRs by reverse transcriptase (RT), followed by integration into the host genome by integrase (1). By virtue of this copy-and-paste mechanism, retrotransposons are thought to have efficiently invaded eukaryotic genomes. Being a large part of eukaryotic genomes, these LTR-retroelements are viewed as major players in eukaryote evolution (2).
A large number of studies on the early replication steps of lentiviruses and gammaretroviruses, such as HIV-1 and MoMuLV, respectively, have been carried out in in vitro reconstituted systems and in cell culture [reviewed in (3–5)]. Collectively the findings show that reverse transcription occurs within the virion nucleocapsid (NC) structure formed of the genomic RNA coated by molecules of NC protein, and starts by NC-mediated annealing of a specific cellular primer tRNA to a unique 5′ genomic primer binding site (PBS), followed by RT-directed cDNA synthesis during which RT is assisted by NC (3–5). Much less studies have been carried out on the early replication steps of ancient retrotransposons, such as TYs of yeast. Although a similar basic mechanism appears to operate, such as RT-directed cDNA synthesis by extension of a specific cellular tRNA annealed to a genomic PBS within a ribonucleoprotein structure, several important differences were discovered. The genomic PBS is not unique but multipartite in TY1 and TY3, and unique genomic 5′–3′ interactions appear to exert a control over reverse transcription and consequently on the genetic amplification of these retrotransposons (6–8).
As it is well documented for HIV-1, NC protein in its mature form plays critical roles in the conversion of the genomic RNA into proviral DNA by RT through specific and tight interactions with the genomic RNA, primer tRNALys,3, RT and the newly made cDNA [reviewed in (3–5)]. Interestingly, retrotransposon NC proteins can either contain a canonical CCHC zinc finger RNA-binding motif and be processed by protease cleavage of the Gag polyprotein, such as in TY3, or else Gag is not processed and does not contain a zinc finger motif, such as in TY1 (9–11). Nevertheless, the retroviral NC functions appear to be conserved in these retrotransposons since the C-terminal region of TY1 Gag chaperones the annealing of primer tRNAMet,i to the 5′ multipartite PBS, mediates TY1 RNA dimerization, and assists cDNA synthesis by the homologous RT (12).
Gypsy is a retroelement present in the germ line of the fruit fly Drosophila melanogaster and can spread via cell-free viral infection. Thus, Gypsy can be considered both as an active retrotransposon and an infectious retrovirus (13,14). In agreement with this notion, the genetic structure of Gypsy is similar to that of the murine gammaretrovirus MuLV with Gag, Pol and Env flanked by two LTRs (15,16). However, the Gypsy Gag structural protein is not processed into Matrix, Capsid and NC proteins (B.V. Syomin and A. Pelisson, unpublished data), which is reminiscent of Gag of the yeast TY1.
These functional and genetic features of Gypsy make it a very attractive model to study replication of a mobile genetic element, which is at the frontier between ancient retrotransposons and retroviruses. To that end we set up an in vitro replication system using Gypsy RNAs representing the genomic RNA 5′ and 3′ regions, and cellular primer tRNALys,2, and investigated their interactions with a putative NC-like domain in Gag. Here we report that an unstructured domain of Gypsy Gag has the hallmarks of an active retroviral NC. This NC-like domain forms ribonucleoparticle-like complexes upon binding Gypsy RNA in vitro. In addition, two distinct primer tRNALys,2 binding sites of 11 nt were identified at the 5′ and 3′ ends of the genome. We also found that the Gypsy NC-like peptide can direct the annealing of tRNALys,2 to these Gypsy PBSs but only the 5′ PBS appears to be indispensable for cDNA synthesis in vitro and in Drosophila cells.
MATERIALS AND METHODS
Plasmid construction
Template DNA pBSG8
The XhoI fragment (−39 to 6949) of Gypsy DNA (GenBank accession no. M12927) was inserted into the SalI site of the Bluescribe F′. The clone was selected so that the EcoRI site of the polylinker was at the 5′ end.
Plasmid DNAs encoding Gypsy 5′ and 3′ RNAs were constructed by cloning a high fidelity PCR generated fragment containing two restriction sites at its ends, using the pBSG8 clone DNA as template and the oligonucleotide primers described in Supplementary Table 1.
The tRNALys,2 encoding DNA was constructed as described previously (17).
Wild-type plasmid DNA used for transfection
Wild-type constructs pDm111 (15) and 111xw (Nathalie, unpublished data) were kindly provided by N. Lyubomirskaya. They both contain the same functional Gypsy/mdg4 element except that, in 111xw, a PvuI site replaced the XhoI restriction site of the 3′ LTR. The EcoRI–PstI fragment containing the 816 3′ nt of Gypsy, including the modified 3′ LTR, was subcloned into pBluescript II KS(-) (Stratagene) to produce clone DNA #p18. The LTR+130UTR plasmid (18), kindly provided by S. Jensen, was used as a template to PCR amplify the 130 bp long copia enhancer with the following KpnI and XhoI primers: 5′-GGGGTACCCAGTCCATGCCTAATAAAC-3′ and 5′-ACCGCTCGAGCTGAGAAGGAAATAATTTC-3′.
The amplified DNA fragment was cut with KpnI and XhoI and ligated to the 5′ end of the 6.4 kb XhoI–EcoRI Gypsy fragment of pDm111 into the pBluescript II KS(-) vector, yielding construct DNA #p19. The 800 bp EcoRI–BamHI fragment from DNA #p18 was inserted into DNA #p19, giving rise to the full-length wild-type (WT) Gypsy construct.
Mutant plasmid DNAs used for transfection
A frameshift starting at codon 40 of the Gag gene was obtained by filling in with Klenow polymerase the NcoI site of the WT plasmid. The 1.2 kb XhoI–KpnI fragment from WT was subcloned into pBluescript II KS(-) to delete the putative 5′ PBS sequence (TGGCGCCCAAC) using the QuikChange® system (Stratagene) with oligonucleotides used for pBSG8-CG2 (Supplementary Table 1). The XhoI–NcoI 1 kb mutated fragment was then excised and swapped with the corresponding wild-type fragment of the WT plasmid, generating the Δ5′ PBS Gypsy DNA. The same procedure was used to delete the four 3′ nt (CAAC) of the 5′ PBS. The 3′ PBS sequence (CCACGACCCTG) was deleted from DNA #p18 using the QuikChange® system (Stratagene) with the oligonucleotides used for pBSG8-CG4 (Supplementary Table 1). The modified EcoRI–BamHI fragment was then inserted into DNA #p19 to yield the Δ3′ PBS Gypsy DNA. All DNA constructs were verified by sequencing.
Proteins and peptides
The TYA1-D NC-like peptide was obtained by opfp chemical synthesis and purified by high-performance liquid chromatography (HPLC) as described before (12,19). The murine leukemia virus (MLV) and HIV-1 NC basic peptides were also obtained by opfp chemical synthesis and purified to homogeneity as described before (19–22). Using the one letter code representation, the MLV-NC peptide is: ′RQGGERRRSQLDRDQCAYCKEKGHWAKDCPRKPRGPRAT′ where the unique zinc finger is underlined, and the HIV-1 NC peptide is: ′TVKCFNCGKEGHIAKNCRAPRKKGCWKCGKEGHQMKDCTERQ′ where the two zinc fingers are underlined. All peptides were dissolved at 1 mg/ml in a buffer containing 30 mM HEPES (pH 6.5), 30 mM NaCl and 0.1 mM ZnCl2. MLV RT was from Invitrogen.
DNA encoding the NC-like region of Gypsy Gag was obtained by PCR amplification using pBSG8 as template (see above) and inserted into pIVEX2.4d (Roche) using the SacII and BamHI sites. The sequence of the Gypsy NC-like peptide is shown in Figure 2B. In addition, a His-tag was present at its N-terminus (MSGSHHHHHHSSGIEGRG). The peptide was expressed in Escherichia coli after overnight induction at 18°C since it proved to be highly toxic at temperatures above 20°C. The peptide was purified in denaturing conditions on Ni-NTA column (Qiagen) and eluted with 100 mM NaH2PO4, 10 mM Tris–HCl, 8 M urea at pH 4.5. Fractions containing the peptide with the highest degree of purity were further diluted with 3 vol of 30 mM HEPES (pH 6.5), 30 mM NaCl and 0.1 mM ZnCl2 and chromatographed on an Amicon ultra-4 column. The peptide was stored at 1 mg/ml in the above buffer. Note that all buffers were extensively degassed before use.
DNA templates and RNA synthesis
Plasmid DNAs for 5′ RNA synthesis were generated by digesting pBSG8-CG1 (wt) and pBSG8-CG2 (ΔPBS) with XmnI (position 433). DNAs for 3′ RNA and 5′–3′ RNA synthesis were obtained by digesting pBSG8-CG3 to CG8 (wt and mutants) with NsiI (position 6941), treated by the Klenow polymerase to remove the 3′ strand overhang before in vitro transcription.
DNA template for tRNALys,2 synthesis was obtained by digesting the tRNALys,2 plasmid with BstNI in order to generate the primer tRNA ending at CCA.
All RNAs were prepared by in vitro transcription with T7 RNA polymerase according to the manufacturer's instructions (Promega). Gypsy RNAs were further purified by spin-column chromatography (S-300 HR, Amersham Biosciences) and dissolved at 1 mg/ml in sterile water.
Primer tRNALys,2 was purified by PAGE in 7 M urea and 0.5× TBE, recovered and dissolved at 0.05 mg/ml, and subsequently heat-denatured and slowly cooled down in the presence of 1 mM MgCl2 for proper folding. Primer tRNALys,2 was labelled by incorporation of [32P]UMP during transcription.
Binding of the Gypsy NC peptide to RNA
32P-labelled Gypsy 5′ or 3′ RNA (5 × 10−8 M) was incubated with Gypsy NC, TYA1-D, MLV-NC or HIV-1 NC at protein to nucleotide molar ratios as indicated in the figure legend, at 30°C for 5 min in 10 μl assays containing 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 0.1 mM MgCl2, 5 mM DTT, 0.01 mM ZnCl2 and 8 U RNasin (Promega). Reactions were stopped by adding 5 mM EDTA and centrifuged for 30 min at 13 000 r.p.m. at 4°C. Supernatants were completely removed by pipetting. Subsequently, 32P radioactivity in the supernatant and pellet fractions was monitored in a Packard 1900 TR scintillation counter.
Primer tRNALys,2 annealing to Gypsy RNA
Gypsy RNA, wt or mutant (5 × 10−8 M), and 32P-tRNALys,2 (6 × 10−8 M) were incubated with nucleic acid chaperone (at concentrations indicated in figure legends) at 30°C for 5 min in 10 μl assays containing 20 mM Tris–HCl (pH 7.5), 30 mM NaCl, 0.1 mM MgCl2, 5 mM DTT, 0.01 mM ZnCl2 and 8 U RNasin (Promega). Reactions were stopped by addition of SDS/EDTA (0.5%/5 mM final concentrations) and proteins were subsequently removed by proteinase K digestion (3 μg) at room temperature for 10 min and RNAs were phenol–chloroform extracted. RNAs were analysed by 1.3% native agarose gel electrophoresis in 0.5× TBE. The gel was fixed with 10% trichloroacetic acid, dried and autoradiographed. 5′ 32P-labelled FX174 DNA HinfI markers (Promega) were used for size determination (data not shown). Quantifications were carried out by laser scanning as described before (8).
Synthesis of Gypsy cDNA in vitro
Gypsy 5′ or 5′–3′ RNA (5 × 10−8 M) and 32P-labelled tRNALys,2 (1 × 10−7 M) were incubated with nucleic acid chaperone (at protein concentrations indicated in the figure legend) at room temperature for 10 min in 10 μl buffer containing 20 mM Tris–HCl (pH 7.0), 30 mM NaCl, 0.1 mM MgCl2, 5 mM DTT, 0.01 mM ZnCl2 and 10 U RNasin (Promega) (20–22).
The reaction volume was then increased to 25 μl by addition of 100 U of MLV RT (Invitrogen), 0.25 mM of each dNTP, 30 mM NaCl and 2.8 mM MgCl2. Incubation was for 5 min at room temperature, and then 1 h at 30°C to allow synthesis of Gypsy minus strand cDNA. Reactions were stopped by adding SDS/EDTA (0.5%/5 mM final concentrations) and heated for 3 min at 60°C. Proteins were removed by phenol–chloroform extraction and nucleic acids were precipitated by ethanol. Samples were resuspended in 10 μl formamide buffer (97% formamide, 1 mM EDTA, 0.02% bromophenol blue and 0.02% xylene cyanol), denatured for 2 min at 95°C and resolved by denaturing 8% PAGE-7 M urea in 0.5× TBE. Subsequently the gel was fixed, dried and autoradiographed. 5′ 32P-labelled FX174 DNA HinfI markers (Promega) were used for size determination.
Inhibition of self-primed cDNA synthesis in vitro
Gypsy 5′ RNA (5 × 10−8 M) was incubated with the NC-like peptide as described above but in the absence of the tRNA primer. For cDNA synthesis, the reaction volume was increased to 25 μl by addition of 100 U of MLV RT (Invitrogen), 30 mM NaCl and 2.8 mM MgCl2, and nucleotides (0.25 mM dATP, dGTP, dTTP, 25 μM dCTP and 20 μCi [32P]dCTP). Incubation, product purification and gel analysis were the same as for initiation of cDNA synthesis by primer tRNA (see section above).
DNA transfection into Drosophila cells and Southern blotting
Exponentially growing Drosophila hydei Dh33 adherent cells were grown at 26°C in Schneider medium supplemented with 10% fetal calf serum (FCS). About 3 × 106 cells were seeded in 9.6 cm2 wells containing 2 ml medium. They were transfected 24 h later with 0.8 μg of plasmid DNA, according to the protocol recommended by Qiagen: 100 μl DNA in EC buffer, 6.4 μl enhancer solution, 20 μl Effectene and 600 μl culture supernatant were successively mixed at 10 min intervals and deposited on the remaining 1.4 ml of culture. Forty-eight hours later, 2 ml of medium were added to the cells, which were transferred into a 25 cm2 flask. Three days after transfection with the LTR+130UTR control plasmid (18), up to 10% of the cells scored positive for X-Gal staining.
Transfected cells were collected at 72 h, then washed twice with 8 ml PBS and the pellet was resuspended in 200 μl [10 mM Tris–HCl (pH 8), 1 mM EDTA and 10 μg·ml−1 RNase I). Lysis was achieved by adding 200 μl [200 mM Tris–HCl (pH 8), 200 mM EDTA and 2% SDS] and the mixture was incubated for 30 min at 70°C. Proteins were precipitated by addition of 60 μl 3 M KoAc (pH 5.2) and incubating the mixture for 30 min at 0°C. The supernatant was successively extracted with phenol/chloroform (1:1) and chloroform/isoamyl alcohol (24:1) and adding 0.3 M sodium acetate (pH 5.2) and 70% ethanol precipitated the DNA.
Half of each extract was digested with 60 U of the methylation-sensitive DpnI restriction enzyme (New England Biolabs), ethanol precipitated and then split into four parts, which were incubated with restriction enzymes as indicated in Figure 9.
Identical amounts of nuclease restricted DNA were run on a 0.7% agarose gel in TBE 1×, partially depurinated, denatured and capillary transferred in 10× SSC on a Nytran N membrane (Schleicher & Schuell). Hybridization with a randomly primed DNA probe—Gypsy coordinates 3097–5759 (16)—was performed in 50% formamide, 5× SSC, 5× Denhardt's solution, 0.1% SDS, at 42°C. After washing at 55°C in (0.1× SSC and 0.5% SDS), the membrane was exposed in a Storm 820 phosphorimager (Molecular Dynamics).
RESULTS
Identification of a NC-like region in Gypsy Gag (ORF1)
The NC proteins of retroviruses and retroelements are small basic proteins with nucleic acid chaperone activities, generated upon processing of the Gag polyprotein by the viral protease (3). Most NC proteins are characterized by the presence of one or two Cx2Cx4Hx4C motifs, called CCHC zinc fingers (23), constituting a specific RNA-binding motif (4,24). However, a number of LTR-containing retroelements—including Gypsy—lack zinc fingers and their Gag is not processed by the protease into matrix, capsid and NC proteins [(12,25,26), and B.V. Syomin and A. Pelisson, unpublished data]. Nevertheless, NC functions may be conserved even in these cases, as evidenced by the Gag protein of the yeast TY1 retrotransposon. Indeed the Gag C-terminal domain of TY1 possesses nucleic acid binding and chaperoning properties, promoting replication primer tRNAMet,i annealing to the RNA genome and initiation of cDNA synthesis by RT (12).
Since NC activity appears to be ubiquitous in functional retroelements (27), we decided to search for an NC-like domain in Gypsy Gag (ORF1). Owing to the absence of a consensus RNA-binding motif or considerable homology to known NC proteins [(26) and data not shown], we took advantage of physico-chemical features probably shared by all NC and NC-like proteins.
The regions flanking the zinc fingers in NC proteins contain low amino acid complexity regions and are rich in proline and basic amino acid residues (26). Proline and charged amino acids as well as low complexity were suggested to contribute to the flexibility of a polypeptide chain (28). Indeed, the solution structures of NCp7 and NCp10 show that these proteins—with the exception of the short zinc finger region(s) (see Materials and Methods)—are poorly structured when not bound to nucleic acids (29–31). This flexibility (i.e. intrinsic disorder) might be an important prerequisite both for the multimerization of NC proteins (32) and for their RNA chaperone function (33,34). Based on these considerations, we propose that the presence of a markedly basic, disordered region in retroviral/retroelement Gag may be a good indicator of NC function in the absence of well characterized RNA-binding domain(s) with CCHC zinc fingers.
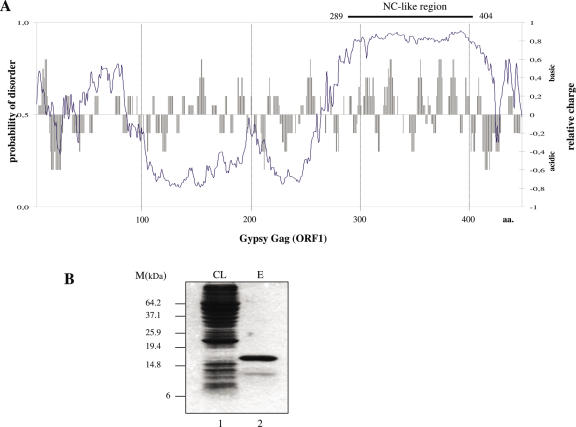
Relatively long unstructured regions can be predicted from amino acid sequence with considerable accuracy. We used the DisProt VL3-H predictor [http://www.ist.temple.edu/disprot/predictor.php, (35)] to assess the probability of disorder in Gypsy Gag. As shown in Figure 1A, the N-terminal and central regions of Gag are predicted to be mostly ordered and do not show a distinctive basic character. However, an approximately 100 amino acid region at the C-terminal end of Gag (between 300 and 400 amino acid) contains predominantly basic amino acid clusters in a putatively disordered environment, a pattern characteristic of retroviral NC proteins [(34) and data not shown]. In agreement with this prediction, an arginine-rich region (355–389 amino acid) has been previously proposed to act as a possible RNA-binding domain of Gypsy Gag (36).
Figure 1.
Identification of the NC-like region in Gypsy Gag. (A) Putative NC-like region of Gypsy Gag. The bar chart illustrates the charge distribution of amino acids in Gypsy Gag (ORF1), as calculated by the charge function of the EMBOSS package, using default parameters and a sliding window of 5 amino acids. Computer prediction of disordered regions (solid line) was obtained using the DisProt VL3-H predictor (http://www.ist.temple.edu/disprot/predictor.php) (35). An amino acid with a disorder score above or equal to 0.5 is considered to be in a disordered environment, while below 0.5 in an ordered environment. (B) Expression and purification of the NC-like peptide. The C-terminal part of ORF1 contains several basic amino acid clusters and is predicted to be disordered in its unliganded state, a common feature of retroelement NC proteins (see text for explanation). This region encompassing 289–404 amino acids was amplified and cloned into the pIVEX2.4d vector. The peptide was amplified in E.coli and obtained in a purified form (see Materials and Methods). CL is the clear E.coli lysate (lane 1) and E the purified Gypsy NC-like peptide (lane 2). Note that the peptide was expressed only at a very low level in the CL. Purification steps allowed us to obtain a peptide more than 95% pure (lane 2). The minor protein band corresponds to an N-terminal cleavage product of the Gypsy NC-like peptide.
The Gag region encompassing residues 289–404 was PCR-amplified and cloned into the pIVEX2.4d vector. The corresponding peptide was found to be toxic in E.coli and to aggregate (data not shown). To circumvent these difficulties it was necessary to express the peptide at 18°C. Due to a very low level of expression (Figure 1B, lane 1) we had to insert a His-tag at the N-terminus in order to achieve an efficient purification and to obtain a peptide at about 95% purity (Figure 1B, lane 2). This Gypsy Gag peptide was found to bind Gypsy RNA and tRNA with a high affinity similar to the NC-like peptide derived from the yeast retrotransposon TY1 Gag, termed TYA1-D (see Figure 2) (data not shown). This prompted us to investigate its RNA condensing and chaperoning properties using in vitro generated RNAs mimicking the 5′ and 3′ regions of the Gypsy genome (Figure 3).
Figure 2.
The Gypsy NC and TYA1-D peptides. Amino acid sequences of the NC-like peptides of the yeast retrotransposon TY1 (TYA1-D) and Gypsy NC are shown according to the one letter code. Note the presence of a large number of basic residues and histidines, and the absence of a canonical ‘CCHC’ Zinc finger motif as in vertebrate retroviruses and in TY3 NCp9 (26). Both peptides were found to be basic and disordered according to computer predictions (see Figure 1A).
Figure 3.
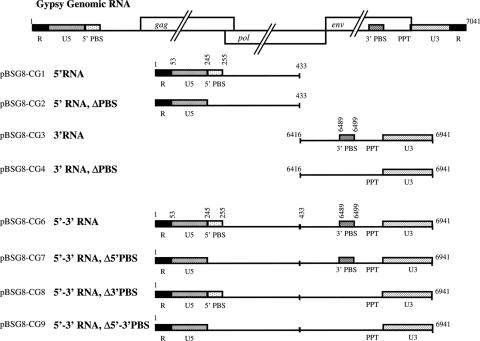
Gypsy RNAs generated in vitro. Schematic representation of the Gypsy genomic RNA where the U3, R and U5 regions of the LTR, the 5′ and 3′ PBS and the Gag, Pol and Env open reading frames are depicted. PBS stands for primer tRNA-binding site and PPT for polypurine tract. The Gypsy 5′ RNA (positions 1 to 433) includes the R repeat, the 3′ region of the LTR (U5) and the 5′ PBS. The Gypsy 3′ RNA (positions 6416 to 6941) encompasses the 3′ PBS and the 5′ region of the LTR (U3). The recombinant 5′–3′ Gypsy RNA contains the above 5′ and 3′ RNA sequences joined to form a single RNA. Numbering is with respect to the genomic RNA positions. RNAs with the PBS mutated to a 6 nt SpeI site (Δ5′ PBS) or to a 6 nt EcoRV site (Δ3′ PBS) are depicted according to the same representation. See table for the cloning strategies and oligonucleotides used to that end (Supplementary Table 1).
The Gypsy NC peptide has RNA-binding and condensing properties
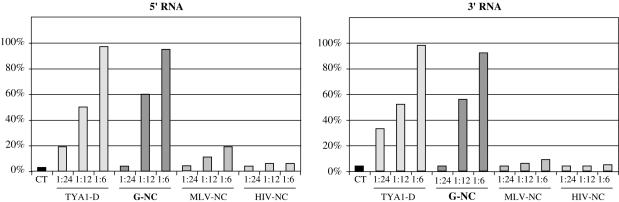
One hallmark of viral RNA chaperone proteins, such as retroviral NC proteins is that they cause RNA aggregation and condensation (12). Under in vitro conditions, large ribonucleoprotein complexes are formed and can be recovered by centrifugation. To analyse the ability of Gypsy NC to form large ribonucleoproteins, Gypsy NC–32P-RNA complexes were formed at 30°C and recovered by centrifugation at 13 000 r.p.m. (see Materials and Methods). Radioactivity in the supernatant and in the nucleoprotein-containing pellet was monitored. Results show that the Gypsy NC (G-NC) caused RNA condensation at a protein to nucleotide molar ratio of 1:6, both with the Gypsy 5′ and 3′ RNAs (Figure 4). At a protein to nt molar ratio of 1:12, 50–60% of the RNA was found in nucleoprotein complexes (Figure 4) in agreement with the fact that about half of the Gypsy 32P-RNA was bound to Gypsy NC as indicated by gel retardation assays (data not shown). The TYA1-D peptide (12) was used as a positive control and, as expected, had a similar RNA condensing activity (Figure 4).
Figure 4.
Binding of the Gypsy NC-like peptide to Gypsy 5′ and 3′ RNAs. Gypsy NC-like peptide and 32P-labelled 5′ and 3′ RNAs were synthesized as described in Materials and Methods. Binding of Gypsy NC (G-NC) was monitored by gel retardation (data not shown) and nucleoprotein complex formation. Complexes were recovered by centrifugation as reported in methods. The same experiments were carried out with the TYA1-D peptide and with MLV and HIV-NC peptides (see Materials and Methods for peptide sequences). Optimal complex formation with the G-NC and TYA1-D peptides was found to take place at a protein to nt molar ratio of 1 to 6, as previously observed for retroviral NC proteins, such as HIV-1 NCp7 and MuLV NCp10 (20,45). Note that the basic HIV-NC and MLV-NC peptides did not form large amounts of ribonucleoprotein complexes (HIV-NC and MLV-NC on both panels); although they bind RNA [see Refs (20–22)]. CT stands for RNA alone.
The observed RNA aggregation cannot be exclusively attributed to charge neutralization caused by the basic amino acids in Gypsy NC (pI = 10.46) and TYA1-D (pI = 10.24), since the similarly basic MLV-NC and HIV-1 NC peptides (with pI values of 10.2 and 9.42, respectively) failed to induce considerable RNA aggregation of the Gypsy 5′ and 3′ RNAs (Figure 4; MLV and HIV-NC bars).
The Gypsy NC peptide directs annealing of tRNALys,2 to two 5′ and 3′ PBSs
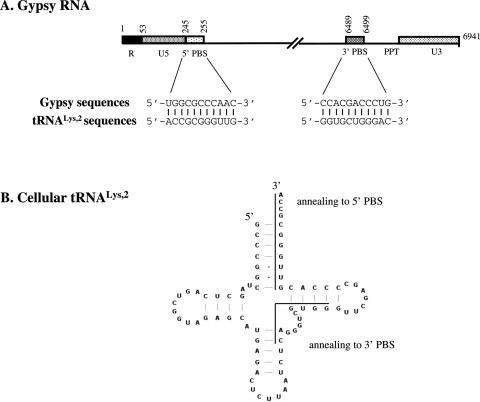
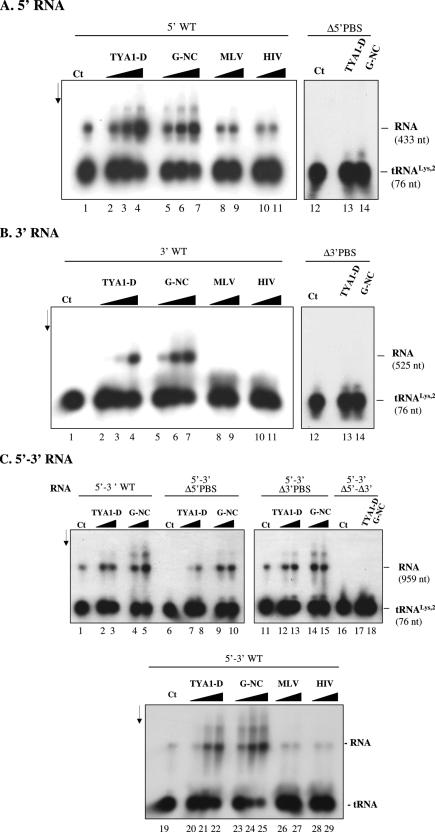
In order to set up an in vitro Gypsy replication system, we needed to know whether the 5′ PBS was unique, bipartite or multipartite as in retroviruses and in the yeast retrotransposons TY3 and TY1, respectively (3,6,7). Sequence alignments using tRNALys,2 and the complete Gypsy sequences were performed. As reported in Figure 5A, a PBS of 11 nt in length and rich in GC is located immediately 3′ of the 5′ LTR at positions 245–255, complementary to the 3′ end of the tRNALys,2 acceptor stem (Figure 5B). Interestingly, another putative tRNALys,2 binding site was found close to the genomic RNA 3′ end at positions 6489 to 6499, also 11 nt in length and rich in GC (Figure 5A). This second PBS is complementary to a different region of tRNALys,2, namely part of the anticodon and TΨC stems (Figure 5B). To assess the role of these 5′ and 3′ PBSs in primer tRNALys,2 annealing and initiation of reverse transcription, we generated several different RNAs mimicking the 5′ and 3′ regions of Gypsy genomic RNA containing the wild-type sequences, or a deleted PBS, and recombinant RNAs where the 5′ and 3′ regions have been fused in the wild-type sequence context or containing either one or both deleted PBSs (Figure 3; see Gypsy RNAs named BSG8-CG 1 to 9).
Figure 5.
The primer tRNALys,2 binding sites on the Gypsy genomic RNA. (A) Gypsy RNA: the 5′ and 3′ regions of the Gypsy genomic RNA are shown. Sequence complementarities of 11 nt between 5′ and 3′ PBS and replication primer tRNALys,2 are indicated. (B) Secondary structure of cellular primer tRNALys,2: sequences possibly involved in tRNA annealing to the 5′ and 3′ PBSs of Gypsy RNA are indicated by lines.
To examine primer tRNALys,2 annealing to the 5′ and 3′ PBSs we used the 5′, 3′ and 5′–3′ Gypsy RNAs generated in vitro (see Materials and Methods, and Figure 3). The Gypsy NC (G-NC) or the TYA1-D peptide was added to the assays together with 32P-labelled tRNA. At the end of the incubation period, 32P-labelled nucleic acids were purified by proteinase K (PK) digestion and phenol extraction, and subsequently analysed by gel electrophoresis in native conditions followed by autoradiography. As reported in Figure 6A, the Gypsy NC and TYA1-D peptides promoted extensive annealing of tRNALys,2 to the 5′ PBS (lanes 2–7) and, as expected, not to the 5′ RNA lacking the PBS (lanes 13–14). Both peptides also chaperoned the annealing of tRNALys,2 to the 3′ PBS, but somewhat less efficiently than to the 5′ PBS (Figure 6B, lanes 2–7). Despite many attempts, the level of tRNALys,2 annealing to the 3′ PBS never reached more than 30–40% with TYA1-D and 40–60% with Gypsy NC as compared with the 5′ PBS. As expected, in the absence of 3′ PBS no tRNA annealing took place (lanes 13–14). At the same time, the MLV and HIV-NC peptides were found to be poorly active (Figure 6A and B, lanes 8–11) although they contain zinc fingers shown to specify direct NC/RNA recognition (3,5,24).
Figure 6.
Hybridization of primer tRNALys,2 to the Gypsy 5′ and 3′ PBSs. Gypsy RNA and 32P-labelled primer tRNALys,2 were incubated at 30°C for 5 min. RNA complexes were purified by SDS-PK treatment and phenol extraction (see Materials and Methods). The nature of the RNA, 5′ wt, Δ5′ PBS, 3′ wt, Δ3′ PBS, recombinant 5′–3′ (either wt, Δ5′ PBS, Δ3′ PBS or Δ5′–Δ3′ PBS) is indicated at the top of each panel. Panel (A) stands for 5′ RNA, (B) for 3′ RNA and (C) for 5′–3′ RNA. Control was with Gypsy RNA and 32P-labelled tRNALys,2 but without NC protein as shown in lanes 1 and 12 (A and B), and 1 and 19 (C). (A and B): TYA1-D and Gypsy NC (G-NC) were added to the assays at protein to nucleotide molar ratios of 1:30, 1:15 and 1:7 (lanes 2–4 and 5–7, respectively). MLV and HIV-NC peptides were at peptide to nt molar ratios of 1:15 and 1:7 (lanes 8–9 and 10–11, respectively). When the Δ5′ PBS RNA or the Δ3′ PBS RNA was used, the peptide to nt ratio was 1:7 (lanes 13–14). Note that primer tRNALys,2 annealed at a low level to the 5′ RNA without the chaperone [(A) lane 1] and annealing was optimal at protein to nt molar ratios of 1:15 to 1:7 [lanes 3–4 and 6–7 in (A)]. Quantifications made by laser scanning indicated that total percentages of tRNA annealed to the 5′ PBS increased from 15–20 to 55–65% upon addition of TYA1-D and G-NC, but did not change after addition of the MLV and HIV peptides (average values of three independent assays). Primer tRNALys,2 did not anneal to the 3′ RNA without chaperone [(B), lane 1] and annealing was optimal at protein to nt molar ratio 1:7 (lanes 4 and 7). Quantifications made by laser scanning indicated that total percentages of tRNA annealed to the 3′ PBS increased from 0 to 40–50% upon addition of TYA1-D and G-NC, but did not change after addition of the MLV and HIV peptides (average values of three independent assays). (C): TYA1-D and Gypsy NC (G-NC) were added to the assays at protein to nucleotide molar ratios of 1:15 and 1:7 (lanes 2–3, 7–8 and 12–13, and 4–5, 9–10, and 14–15, respectively). For Δ 5′–Δ3′ PBS RNA, ratio was 1:7 (lanes 17–18). Additional experiments with TYA1-D and G-NC were at molar NC to nt ratios of 1:30, 1:15 and 1:7 (lanes 20–22 and 23–25, respectively) and with MLV and HIV peptides ratios were 1:15 and 1:7 (lanes 26–27 and 28–29, respectively). Quantifications made by laser scanning indicated that total percentages of tRNA annealed to the 5′ and 3′ PBSs increased from 8–12 to 40–85% upon addition of TYA1-D and G-NC, but did not change after addition of the MLV and HIV peptides (average values of three independent assays; see also Figure 7). Sizes of tRNA and Gypsy RNAs (in nt) are indicated on the right for 5′ RNA (433 nt), 3′ RNA (525 nt), 5′–3′ RNA (959 nt) and tRNALys,2 (76 nt).
We used a recombinant 5′–3′ RNA representing the 5′ and 3′ regions of the Gypsy genomic RNA to better understand what could be the interactions between the 5′ PBS–tRNA and the 3′ PBS–tRNA complexes since different tRNALys,2 sequences of 11 nt should be base paired to the Gypsy 5′ and 3′ PBSs (Figure 5B), thus possibly forming a link between the ends of Gypsy genomic RNA in a manner similar to yeast TY3 RNA and primer tRNAMet,i (7). Overall, the Gypsy NC and TYA1-D peptides still directed tRNA annealing to the PBSs (Figure 6C, lanes 2–5 and 20–25). Deleting the 5′ or 3′ PBS did not result in levels of tRNA annealing to the 5′ PBS or 3′ PBS (Figure 6C, lanes 7–10 and 12–15, respectively) similar to those obtained with the 3′ RNA or 5′ RNA alone (compare with wt RNAs in Figure 6A and B). As before, the MLV and HIV-NC peptides were inactive (lanes 26–29). Taken together, these results indicate that complex interactions are probably taking place between Gypsy 5′ and 3′ terminal regions, probably competing with the binding to tRNALys,2.
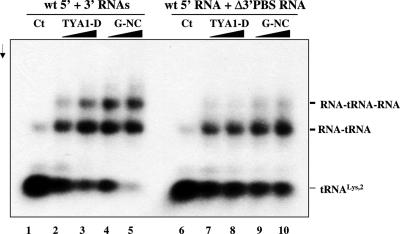
To investigate how tRNALys,2 could possibly establish a bridge between the 5′ and 3′ ends of the Gypsy genomic RNA, we analysed the simultaneous binding of tRNA to the PBSs situated on two RNA molecules. To this end, Gypsy 5′ and 3′ RNAs were co-incubated with tRNA and the TYA1-D or Gypsy NC peptide. After peptide removal by SDS-proteinase K digestion and phenol extraction, RNAs were subjected to electrophoresis in native conditions. As shown in Figure 7, an RNA complex formed of the 5′ and 3′ RNAs, and tRNA was generated upon addition of TYA1-D or G-NC in a dose dependent manner (lanes 2–5) when both PBSs were present. However, this complex did not form upon deleting the PBS from the 3′ RNA (lanes 7–10). Similar results were obtained with the Gypsy 5′ RNA with a deleted PBS (data not shown). These results indicate that the Gypsy primer tRNA can establish a bridge between the 5′ and 3′ ends of the genomic RNA (see Discussion).
Figure 7.
Primer tRNALys,2 bridges the 5′ and 3′ RNA ends. Gypsy 5′ and 3′ RNAs and 32P-labelled primer tRNALys,2 were incubated together at 30°C for 5 min. RNA complexes were purified by SDS-PK treatment and phenol extraction (see Materials and Methods). The nature of the RNA, 5′ wt, 3′ wt and Δ3′ PBS RNA is indicated at the top. Control Gypsy RNAs with 32P-tRNALys,2 but without protein are shown in lanes 1 and 6. TYA1-D or Gypsy NC (G-NC) peptide was added to the assays at protein to nucleotide molar ratios of 1:15 and 1:7 as shown in lanes 2–3, 4–5, 7–8 and 9–10, respectively. Arrow is direction of electrophoresis. Primer tRNALys,2 annealed at a low level to the RNA without chaperone (lanes 1 and 6) and this was optimal at a molar ratio of 1:7 (lane 5). Note that annealing of tRNALys,2 to both 5′ and 3′ PBS caused the formation of a complex made of 5′ and 3′ Gypsy RNAs and tRNA (lanes 4–5) but not when tRNA could not anneal to the 3′ RNA (lanes 9–10).
Influence of the Gypsy NC peptide on cDNA synthesis in vitro
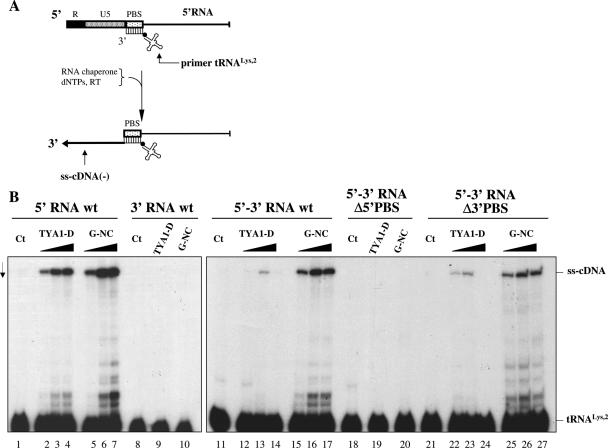
To examine the influence of the Gypsy NC peptide and the role of the 5′ and 3′ PBSs on Gypsy cDNA synthesis, we used the 5′, 3′ and 5′–3′ RNAs as well as the PBS-mutated RNAs (Figure 3). Reverse transcription by the related MLV RT proceeded at 30°C for 1 h, after which cDNAs were recovered by SDS-PK treatment and phenol extraction, and analysed by gel electrophoresis in denaturing conditions (see Materials and Methods). The Gypsy NC and TYA1-D peptides directed tRNALys,2 hybridization to the 5′ PBS (Figures 6 and 7) that allowed strong stop cDNA (ss-cDNA) synthesis by RT using the 5′ RNA template (Figure 8, lanes 2–7). In the absence of NC peptide no ss-cDNA was made, in agreement with the fact that little or no primer tRNA was annealed to the 5′ PBS (lane 1). Since tRNA annealing to the 3′ PBS did not involve its 3′ terminal nucleotides (Figure 5), no cDNA was synthesized using the 3′ RNA (lanes 9–10).
Figure 8.
Role of the Gypsy NC peptide in cDNA synthesis in vitro. (A) Schematic representation of the initiation of reverse transcription on Gypsy 5′ RNA. Reverse transcription of Gypsy 5′ U5 and R RNA sequences leads to the synthesis of the so-called minus strand strong stop cDNA, ss-cDNA(−), by RT extension of primer tRNALys,2. (B) Gypsy 5′ RNA, 3′ RNA or recombinant 5′–3′ RNA and 32P-tRNALys,2 were incubated with or without TYA1-D or Gypsy NC peptide. MLV RT was added together with dNTPs to allow reverse transcription. Assays were processed as described in Materials and Methods and ss-cDNA(−) was denatured and analysed by 10% PAGE in 7 M urea. Controls without protein are shown in lanes 1, 8, 11, 18 and 21. Protein to RNA nucleotide molar ratios were 1:48 (lanes 2, 5, 12, 15, 22 and 25), 1:24 (lanes 3, 6, 13, 16, 23 and 26) and 1:12 (lanes 4, 7, 9, 10, 14, 17, 19, 20, 24 and 27), corresponding to 1.25 × 10−7, 2.5 × 10−7 and 5 × 10−7 M for 5′ RNA and 2.5 × 10−7, 5 × 10−7 and 10−6 M for 5′–3′ RNA. Note that hybridization of tRNALys,2 to the 3′ PBS caused an inhibition of the initiation of Gypsy reverse transcription which was severe with the TYA1-D peptide (compare lanes 2–4 with 12–14) and moderate with the homologous peptide (compare lanes 5–7 with 15–17).
We next examined the influence of the 3′ PBS on ss-cDNA synthesis initiated at the 5′ PBS using the 5′–3′ RNA template. Results were very similar to the previous ones since only the 5′ PBS was capable of directing ss-cDNA synthesis by RT (compare lanes 15–17 and 20) with only a little influence of the 3′ PBS (compare lanes 15–17 and 25–27). However, only the Gypsy NC peptide was effective at stimulating cDNA synthesis using the 5′–3′ RNA template (compare lanes 12–14 and 15–17). Similar results were obtained upon mixing the 5′ and 3′ RNA templates (data not shown). Both the TYA1-D and Gypsy NC peptides were found to strongly inhibit cDNA synthesis by self-priming (Supplementary Figure 1), which further support the specific role of NC protein in proviral DNA synthesis in most, if not all retroviruses (3).
Analysis of Gypsy cDNA synthesis in D.hydei cells
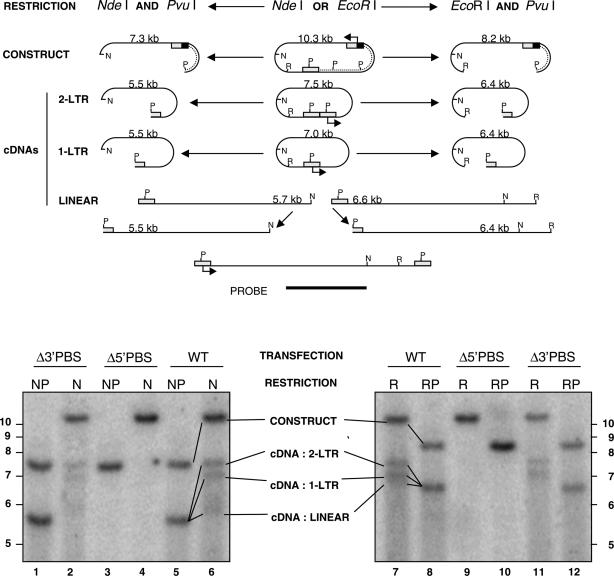
Lyubomirskaya et al. (37) have developed a simple ex vivo assay to look for Gypsy cDNA synthesis, which allows the direct detection of Gypsy reverse transcription by Southern blotting in stably transformed Dh14 cells. These D.hydei cells were chosen because their genome does not cross-hybridize with the D.melanogaster Gypsy sequences. The rationale of this assay is that a small 5′ end deletion in a LTR retroelement still allows the production of a full-length transcript, which is subsequently used as a template to generate a complete cDNA with a restriction map different from that of the deleted donor DNA. This assay benefited here from a slight modification of the donor plasmid where a strong enhancer sequence from the copia retrotransposon (18) was inserted upstream of the Gypsy promoter, replacing the U3 sequence from the 5′ LTR (see Materials and Methods). To improve the signal-to-noise ratio, we took advantage of the fact that the donor plasmid DNA was of prokaryotic origin and could be selectively degraded into small DNA fragments by the DpnI enzyme. The newly made cDNA and a minority of the parental DNA, which happened to have replicated in these Dh cells, are devoid of the methylation tags and are therefore resistant to DpnI.
The results reported in Figure 9 (lanes 5–8, WT) show that the unintegrated cDNA products, i.e. the linear and both the 1-LTR and 2-LTR circles, were found by Southern blotting as early as three days after transfection. To investigate the role of the 5′ PBS we constructed a Δ5′ PBS mutant producing a genomic RNA missing the 11 nt, which hybridize to the 3′ terminus of primer tRNALys,2. Using Gypsy anti-Gag and anti-Env antibodies, we showed by western immuno-blotting of cellular protein extracts that this 5′ PBS deletion did not affect Gypsy protein expression (data not shown). Results show that only the mutant donor plasmid DNA was detected by Southern blotting, clearly indicating that the 5′ PBS is required for Gypsy cDNA synthesis. This is also in complete agreement with the in vitro data (Figure 8, lanes 17 and 20). A smaller 5′ PBS deletion has been made (see Materials and Methods) and results confirmed that the 5′ PBS is essential for Gypsy cDNA synthesis ex vivo (data not shown).
Figure 9.
The 5′ PBS is critical for Gypsy cDNA synthesis ex vivo. D.hydei cells were transfected with WT or PBS-mutated Gypsy DNA as reported in materials and methods. The 10.3 kb transfected DNA is shown at the top. The dotted line represents the vector DNA that was used to clone the Gypsy element with a complete 3′ LTR (rectangle with P, a PvuI site) and a shortened 5′ LTR (square) the U3 sequence of which was replaced by a Copia enhancer (black box). Reverse transcription is expected to regenerate the missing sequence, giving rise to the full-length 7.5 kb Gypsy DNA shown at the bottom. The 2-LTR and 1-LTR circular DNA products of 7.5 and 7.0 kb, respectively, are shown. A total of 1 μg of total DNA were digested with either EcoRI (R) or NdeI (N), blotted and probed with the fragment schematized at the bottom. A unique band is expected to correspond to each category of molecules. The only exception corresponds to the integrated proviruses (not represented here), which if randomly inserted at various distances from the EcoRI or NdeI genomic restriction sites, are expected to produce a smear corresponding to a heterogeneous population of molecules larger than 6.6 and 5.7 kb, respectively. In contrast, in double digests involving PvuI (P) and either NdeI (N) or EcoRI (R), all types of cDNA products should merge into a single band, as shown in lanes 1, 5, 8 and 12). Most of the transfected plasmids have run out of the gel after being degraded into small DNA fragments by the methylation-dependent DpnI enzyme; therefore, the upper band only corresponds to the minority of plasmids, which have lost these prokaryotic methylation tags. Δ5′ and Δ3′ PBS are mutants which only differ from the wild-type Gypsy sequence (WT) by the ΔPBS mutations described in Figure 3, at positions (proviral DNA sequence) 482–492 and 6727–6737, respectively (16). Note that the Δ5′ PBS, but not the Δ3′ PBS deletion resulted in the absence of cDNA synthesis (lanes 3–4 and 9–10).
Next, we constructed the Δ3′ PBS mutant by deleting nt 6727–6737 (Materials and Methods). This Δ3′ PBS mutant displayed a wild-type level of cDNA made (compare lanes 1–2 with 5–6, and 11–12 with 7–8) despite the fact that it does not produce any envelope protein due to the frameshift generated by this 11 nt deletion at the Env 3′ end (data not shown). To further address the role of Gag in the synthesis of Gypsy cDNA, we inserted a frameshift mutation at the 5′ end of Gag. This Gypsy mutant that cannot express the Gag protein was totally unable to direct cDNA synthesis (data not shown). In conclusion, these results confirm the in vitro findings (Figure 8; lanes 17 and 27) and show that the 3′ PBS is not required for cDNA synthesis, at least at this stage of Gypsy replication.
DISCUSSION
The D.melanogaster Gypsy retroelement has a genetic organization similar to that of the widespread vertebrate gammaretroviruses, such as the MLV. In fact the provirus form of Gypsy contains the three canonical open reading frames gag, pol and env flanked by two LTRs. As an infectious enveloped retrovirus, Gypsy can efficiently invade the germ line of permissive flies, upon infection of naive females (38,39). These new Gypsy proviruses can then increase their number by expression in the somatic cells of permissive flamenco females followed by recurrent invasion of the germ line by a process, which, in contrast to virus infection, does not need env (40). Thus, Gypsy can behave both as an infectious retrovirus and as an active retrotransposon (13,14).
A high level of Gypsy transposition might be deleterious for the host genome due to insertional mutagenesis, especially because Gypsy carries a strong DNA insulator (41), which can disrupt the activity of the genes, into which it is integrated by blocking the interactions of distal enhancers with the target promoter. Thus, the inability of Gypsy to extensively invade the genome is conditioned by the cell capacity to repress this retroelement. This control results in reduced steady state levels of Gypsy transcripts (42,43). Moreover, it has recently been shown that synthesis of Gypsy Gag is strictly controlled at the translational level (44).
To understand the dual nature of the D.melanogaster Gypsy and the fact that it appears to be at the frontier between retrotransposons of yeast and simple vertebrate retroviruses, we investigated Gypsy replication in vitro and in cell culture. First, we searched for a NC-like region in Gypsy Gag with all the properties of a bona fide retroviral RNA chaperone protein (45) [reviewed in (3,4)]. To this end, we used a computer-assisted search to look for a peptide domain of low complexity, rich in proline and basic residues, and with a predicted disordered state (33–35). Such a peptide domain, named G-NC, was found at the C-terminal end of the Gypsy Gag, showing notable similarities with the NC-like domain of TY1 Gag (Figures 1 and 2). In fact, the Gypsy NC-like peptide was active in the formation of high molecular weight nucleoprotein complexes and in replication primer annealing to the genomic PBS in a manner very similar to that of the C-terminal Gag peptide of yeast TY1 (TYA1-D; Figures 4 and 6). Like other retroviral NC proteins, the G-NC peptide was found to possess general RNA chaperoning properties in vitro, which can promote profound rearrangements of nucleic acid conformation [reviewed in (27,34)]. Along this line, the optimal chaperoning activity of G-NC takes place at a peptide to nucleic acid molar ratio of one peptide per 6 to 10 nt (see Figure 6) as holds true for retroviral NC proteins and NC of the yeast TY3 retrotransposon (27,34,45). Interestingly, NCp9 of TY3 was found to be active in the Gypsy system (data not shown) whereas the basic MLV and HIV-1 NC peptides were poorly active (Figures 4 and 6) although they contain zinc fingers shown to specify direct NC–RNA interactions in HIV and MLV (3,24,29). The primary structure of the Gypsy Gag and the presence of an active RNA chaperoning domain at its C-terminus suggest that the Gypsy Gag resembles more that of an ancient retrotransposon than the canonical Gag of the gammaretrovirus MLV, formed of the well-defined matrix, capsid and NC domains. On the opposite the Gypsy genomic RNA has long structured untranslated regions, the 5′- and 3′-untranslated regions (UTRs) (Figure 3) similar to the MLV genomic RNA. In addition, the Gypsy 5′-UTR directs translation of Gag and Env via specific internal ribosome entry sites (IRES) that resemble the MLV Gag and Env IRESes (44,46,47).
Second, we characterized two distinct PBSs located at the 5′ and 3′ ends of the Gypsy genomic RNA (Figure 5), which are reminiscent of those found in the yeast TY3 retrotransposon (7). The G-NC peptide promoted the annealing of primer tRNALys,2 to both the 5′ and 3′ PBSs (Figure 6). Furthermore, the simultaneous annealing of primer tRNALys,2 to these two PBSs (Figure 7) resulted in the formation of a 5′–3′ RNA complex, indicating that a tRNA bridge can possibly generate a circular Gypsy RNA as proposed for the TY3 RNA (7). However, Gypsy differs from TY3 since the 3′ PBS is not implicated in the process of cDNA synthesis in vitro and ex vivo (Figures 8 and 9). Yet the exact role of the Gypsy 3′ PBS could be at the level of genome maintenance or translation via formation of a circular RNA. This is presently under investigation.
In conclusion, these findings on the NC-like domain of Gypsy Gag and on the existence of two PBSs indicate that Gypsy resembles active retrotransposons and is distinct from canonical simple retroviruses, such as the MLVs and avian leukosis viruses. In agreement with this conclusion, the env-independent reverse transcription of Gypsy in D.hydei cells (Figure 9) is reminiscent of the fact that the efficiency of Gypsy replication in D.melanogaster females was not affected by the frameshift disruption of the env gene (40). On the other hand, the presence of a single 5′ PBS necessary for cDNA synthesis (Figures 8 and 9), of long structured UTRs, of functional IRESes and Env, and the fact that cell-free Gypsy particles are infectious (13,14) indicate that Gypsy should be considered as an ancient simple retrovirus, similar to the gammaretrovirus MLV. To investigate how much the NC functions are conserved from Drosophila Gypsy to the MLV, substituting MLV-NC by the Gypsy NC-like is now in progress using MLV-based vectors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Marc Ruff (Strasbourg) for helpful advice on peptide expression and purification procedures. Thanks are due to Damien Ficheux (IBCP CNRS) for TYA1-D, MLV-NC and HIV-1 NC peptide synthesis and purification. Gag mutant plasmids were generated by Emeline Sarot with the technical assistance of Geneviève Payen-Groschêne. Work supported by CNRS, ANRS and INSERM to J.L.D., by ARC to A.P. and by CNRS to A.B. R.I.N. is a recipient of an ANRS pre-doctoral fellowship. Funding to pay the Open Access publication charges for this article was provided by ANRS (Agence Nationale de recherches sur le SIDA).
Conflict of interest statement. None declared.
REFERENCES
- 1.Boeke J.D., Stoye J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 2.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.Darlix J.L., Lapadat-Tapolsky M., de Rocquigny H., Roques B.P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 4.Darlix J.L., Cristofari G., Rau M., Pechoux C., Berthoux L., Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- 5.Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 6.Friant S., Heyman T., Wilhelm M.L., Wilhelm F.X. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res. 1996;24:441–449. doi: 10.1093/nar/24.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabus C., Ficheux D., Rau M., Keith G., Sandmeyer S., Darlix J.L. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristofari G., Bampi C., Wilhelm M., Wilhelm F.X., Darlix J.L. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchner J., Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J. Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orlinsky K.J., Sandmeyer S.B. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J. Virol. 1994;68:4152–4166. doi: 10.1128/jvi.68.7.4152-4166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merkulov G.V., Swiderek K.M., Brachmann C.B., Boeke J.D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cristofari G., Ficheux D., Darlix J.L. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 13.Kim A., Terzian C., Santamaria P., Pelisson A., Prud'homme N., Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song S.U., Gerasimova T., Kurkulos M., Boeke J.D., Corces V.G. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 15.Bayev A.A., Jr, Lyubomirskaya N.V., Dzhumagaliev E.B., Ananiev E.V., Amiantova I.G., Ilyin Y.V. Structural organization of transposable element mdg4 from Drosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucleic Acids Res. 1984;12:3707–3723. doi: 10.1093/nar/12.8.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marlor R.L., Parkhurst S.M., Corces V.G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barat C., Le Grice S.F.J., Darlix J.L. Interaction of HIV-1 reverse transcriptase with a synthetic form of its replication primer, tRNALys,3. Nucleic Acids Res. 1991;19:751–757. doi: 10.1093/nar/19.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavarec L., Jensen S., Heidmann T. Identification of a strong transcriptional activator for the copia retrotransposon responsible for its differential expression in Drosophila hydei and melanogaster cell lines. Biochem. Biophys. Res. Commun. 1994;203:392–399. doi: 10.1006/bbrc.1994.2195. [DOI] [PubMed] [Google Scholar]
- 19.de Rocquigny H., Ficheux D., Gabus C., Fournie-Zaluski M.C., Darlix J.L., Roques B.P. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;180:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- 20.Barat C., Lullien V., Schatz O., Keith G., Nugeyre M.T., Gruninger-Leitch F., Barre-Sinoussi F., Le Grice S.F., Darlix J.L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darlix J.L., Vincent A., Gabus C., de Rocquigny H., Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. III. 1993;316:763–771. [PubMed] [Google Scholar]
- 22.Allain B., Lapadat-Tapolsky M., Berlioz C., Darlix J.L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Covey S.N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mely Y., Piemont E., Sorinas-Jimeno M., de Rocquigny H., Jullian N., Morellet N., Roques B.P., Gerard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type I nucleocapsid protein zinc fingers and their involvement in heterologous tRNA(Phe) binding: a steady-state and time-resolved fluorescence study. Biophys. J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer B., Bannert H., Darai G., Flugel R.M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J. Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson-Burch B.D., Voytas D.F. Genes of the Pseudoviridae (Ty1/copia retrotransposons) Mol. Biol. Evol. 2002;19:1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 27.Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 28.Dunker A.K., Lawson J.D., Brown C.J., Williams R.M., Romero P., Oh J.S., Oldfield C.J., Campen A.M., Ratliff C.M., Hipps K.W., et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 29.Summers M.F., Henderson L.E., Chance M.R., Bess J.W., Jr, South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C., III, Hare D.R., et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demene H., Jullian N., Morellet N., de Rocquigny H., Cornille F., Maigret B., Roques B.P. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J. Biomol. NMR. 1994;4:153–170. doi: 10.1007/BF00175244. [DOI] [PubMed] [Google Scholar]
- 31.Morellet N., de Rocquigny H., Mely Y., Jullian N., Demene H., Ottmann M., Gerard D., Darlix J.L., Fournie-Zaluski M.C., Roques B.P. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H-NMR. J. Mol. Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 32.Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 33.Tompa P., Csermely P. The role of structural disorder inthe function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 34.Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53:566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 36.Alberola T.M., de Frutos R. Molecular structure of a gypsy element of Drosophila subobscura (gypsyDs) constituting a degenerate form of insect retroviruses. Nucleic Acids Res. 1996;24:914–923. doi: 10.1093/nar/24.5.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyubomirskaya N.V., Avedisov S.N., Surkov S.A., Ilyin Y.V. Two Drosophila retrotransposon gypsy subfamilies differ in ability to produce new DNA copies via reverse transcription in Drosophila cultured cells. Nucleic Acids Res. 1993;21:3265–3268. doi: 10.1093/nar/21.14.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prud'homme N., Gans M., Masson M., Terzian C., Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelisson A., Mejlumian L., Robert V., Terzian C., Bucheton A. Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem. Mol. Biol. 2002;32:1249–1256. doi: 10.1016/s0965-1748(02)00088-7. [DOI] [PubMed] [Google Scholar]
- 40.Chalvet F., Teysset L., Terzian C., Prud'homme N., Santamaria P., Bucheton A., Pelisson A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gdula D.A., Gerasimova T.I., Corces V.G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl Acad. Sci. USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelisson A., Song S.U., Prudhomme N., Smith P.A., Bucheton A., Corces V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarot E., Payen-Groschene G., Bucheton A., Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronfort C., de Breyne S., Sandrin V., Darlix J.L., Ohlmann T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. RNA. 2004;10:504–515. doi: 10.1261/rna.5185604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prats A.C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J.L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berlioz C., Darlix J.-L. An internal ribosomal entry mechanism promotes translation of MuLV gag polyprotein precursors. J. Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deffaud C., Darlix J.-L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 2000;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]