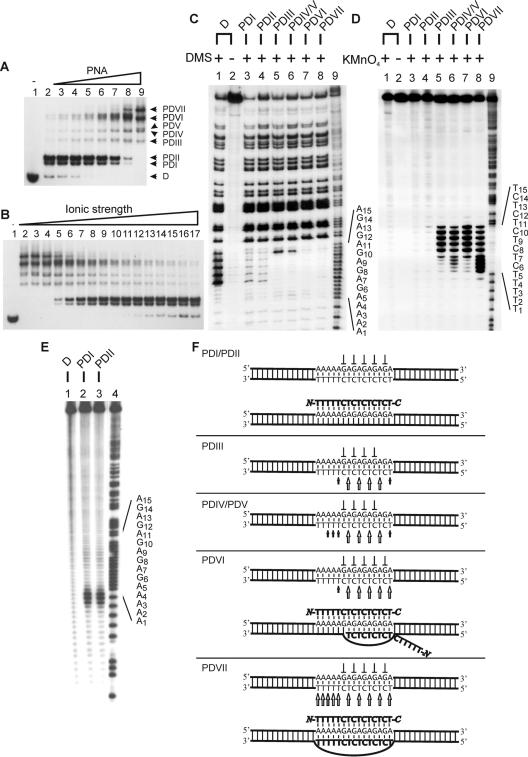
Figure 3.
Binding of PNA pentadecamer to dsDNA (PD15). Autoradiographs showing binding of PNA1940 to dsDNA as investigated by gel-shift analysis (A and B), in situ chemical probing (C and D) and affinity cleavage (E). (A) PNA titration using the following concentrations: lane 1, w/o; lane 2, 0.25 μM; lane 3, 0.5 μM; lane 4, 0.75 μM; lane 5, 1 μM; lane 6, 2 μM; lane 7, 4 μM; and lane 8, 8 μM. Free DNA (D) and the formed complexes PDI15–PDVII15 are indicated. (B) PD15 complex formation as a function of ionic strength. Incubation was carried out using 6 μM PNA and supplemented with KCl as follows: lane 1, w/o; lane 2, 5 mM; lane 3, 10 mM; lane 4, 20 mM; lane 5, 30 mM; lane 6, 40 mM; lane 7, 50 mM; lane 8, 60 mM; lane 9, 70 mM; lane 10, 80 mM; lane 11, 100 mM; lane 12, 120 mM; lane 13, 140 mM; lane 14, 160 mM; lane 15, 180 mM; lane 16, 300 mM; and lane 17, 320 mM. (C) DMS probing of the complexes identified in (A). Lane 1, dsDNA + DMS; lane 2, dsDNA w/o DMS; lane 3, PDI15 + DMS; lane 4, PDII15 + DMS; lane 5, PDIII15 + DMS; lane 6, PDIV15/PDV15 + DMS; lane 7, PDVI15 + DMS; lane 8, PDVII15 + DMS; and lane 9, A/G sequence marker. (D) KMnO4 probing of the complexes identified in (A). Lane 1, dsDNA + KMnO4; lane 2, dsDNA w/o KMnO4; lane 3, PDI15 + KMnO4; lane 4, PDII15 + KMnO4; lane 5, PDIII15 + KMnO4; lane 6, PDIV15/PDV15 + KMnO4; lane 7, PDVI15 + KMnO4; lane 8, PDVII15 + KMnO4; and lane 9, A/G sequence marker. (E) Autoradiograph showing PDI15 and PDII15 subjected to Fe/NTA affinity cleavage. Lane 1, dsDNA + Fe/NTA; lane 2, PDI15 + Fe/NTA; lane 3, PDII15 + Fe/NTA; and lane 4, A/G sequence marker. (F) Structural assignment (arrow, KMnO4 hypersensitivity; perpendicular symbol, DMS protection). The 168 bp Xba–PvuII restriction fragment of p322 was used either 5′-kinase labeled (A–C, E) or 3′-Klenow labeled (D) with 32P-phosphate at the XbaI site. PNA incubations were at pH 6.5 as described in Materials and Methods.