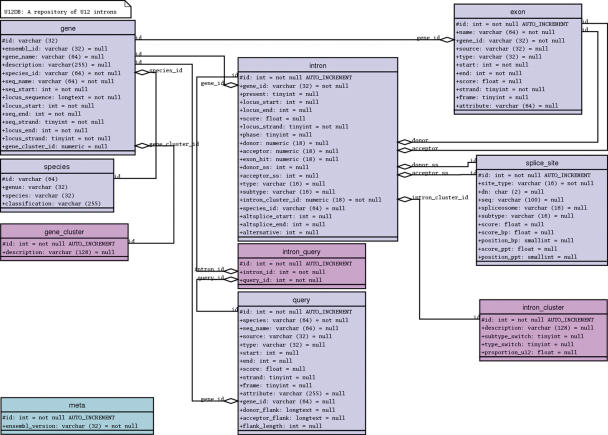
Figure 1.
The U12DB schema. Square rectangles are tables in MySQL. The lines connecting them represent foreign key—primary key relationships. Transcript-confirmed or annotated U12 introns from reference genomes are loaded into the query table. The introns are then associated with Ensembl or TAIR6 genes and these genes are inserted into the gene table. The Ensembl Compara database or a custom-built Inparanoid human/Arabidopsis database is then searched for orthologs. The orthologs are inserted into the gene table and assigned a gene cluster id that is stored in a separate table. Each reference intron is then mapped with Exonerate to each gene in the gene cluster to which its host gene belongs. The introns are assigned an intron cluster id and stored in the intron table. Splice site and exon information are stored in separate tables. The system is designed to be updatable so that intron and gene records are not duplicated but, instead, are updated if they are the target of a new reference intron mapping from the query set. Therefore, there is a many-to-many relationship between query and intron.