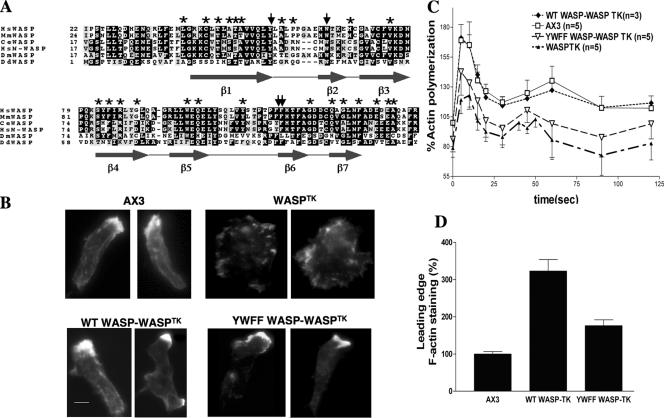
Figure 1.
Conserved WH1 domain aromatic residues are important for WASP function. (A) Comparison of the amino acid sequence of DdWASP WH1 domain with other species: Hs, Homo sapiens; Mm, Mus musculus; Ce, C. elegans; Dm, Drosophila melanogaster; Dd, D. discoideum. Asterisks indicate residues with naturally occurring mutations in WAS patients. Arrows indicate residues mutated to alanine. Predicted secondary structure is indicated below sequence. (B) In vivo F-actin organization as determined by Alexa594-phalloidin staining. WH1 domain mutations (YWFF) reduce F-actin levels at leading edge of WASPTK cells rescued via ectopic WASP expression. Equivalent expression levels of WASP were determined via quantitation of YFP fluorescence and Western analysis. Bar, 5 μm. (C) In vivo actin polymerization assay. Full rescue of WASPTK phenotype by WASP overexpression requires WH1 aromatic residues. (D) Quantitation of F-actin at the leading edge. F-actin levels were determined by measuring the integrated Alexa594-phalloidin signal intensity (area × average intensity) at the leading edge divided by the average intensity of phalloidin staining throughout the cell body. A minimum of 100 cells of each type was taken from at least three independent experiments.