Abstract
Blue light-induced transcription in Neurospora crassa is regulated by the White Collar-1 (WC-1) photoreceptor. We report that residue K14 of histone H3 associated with the light-inducible albino-3 (al-3) promoter becomes transiently acetylated after photoinduction. This acetylation depends on WC-1. The relevance of this chromatin modification was directly evaluated in vivo by construction of a Neurospora strain with a mutated histone H3 gene (hH3K14Q). This strain phenocopies a wc-1 blind mutant and shows a strong reduction of light-induced transcriptional activation of both al-3 and vivid (vvd), another light-inducible gene. We mutated Neurospora GCN Five (ngf-1), which encodes a homologue of the yeast HAT Gcn5p, to generate a strain impaired in H3 K14 acetylation and found that it was defective in photoinduction. Together, our findings reveal a direct link between histone modification and light signaling in Neurospora and contribute to the developing understanding of the molecular mechanisms operating in light-inducible gene activation.
INTRODUCTION
Biochemical signal transduction has been the object of a multitude of studies, mostly devoted to the identification of the molecular components of specific pathways, their epistatic relationships, and their mechanisms of action. Although light signal transduction pathways have been primarily studied in plants (Chen et al., 2004), the filamentous fungus Neurospora crassa has become a preferred eukaryotic model to investigate pathways inducible by blue light. Early biochemical and photobiology studies reported that Neurospora is responsive only to blue light (Sargent and Briggs, 1967; DeFabo et al., 1976). Genes encoding putative red light photoreceptors have been recently discovered in the Neurospora genome (Galagan et al., 2003; Borkovich et al., 2004), but deletion of these genes does not affect any known photoresponses, leaving their function uncertain (Froehlich et al., 2005).
Two proteins are required for blue light perception in N. crassa, White Collar (WC)-1 and WC-2 (Ballario et al., 1996; Linden and Macino, 1997). WC-1 is the photoreceptor (He et al., 2002), with a sensor domain called LOV (for Light, Oxygen, and Voltage), homologous to that of plant phototropins (Huala et al., 1997). WC-1 is associated with WC-2 in vivo, forming a nuclear heterodimer, the White Collar complex (WCC; Talora et al., 1999; Schwerdtfeger and Linden, 2001). At various points in the circadian cycle, other proteins associate with WCC, e.g., the oscillator, Frequency (FRQ; Dunlap and Loros, 2004), and protein kinase C (Franchi et al., 2005), but the two WC proteins are the defining, constant components of this light-sensing complex (Cheng et al., 2002). On blue light irradiation, the conformation of the WCC is thought to change to activate light-dependent genes transiently. The early light-inducible genes, such as albino-3 (al-3) and vivid (vvd), reach their peak induction 15–20 min after a light pulse and are switched off within 1 h (Baima et al., 1991). Initially, WC-1 becomes increasingly phosphorylated and after 1 h the hyperphosphorylated protein begins to turn over (for models, see Talora et al., 1999 and He et al., 2005). As part of a feedback loop, the wc-1 gene itself is subject to light-induced transcription by the WCC (Ballario et al., 1996; Kaldi et al., 2006).
WC-1 and WC-2 are classified as Per Arnt Sim (PAS) transcription factors, which are characterized by a zinc finger-binding domain similar to that of vertebrate GATA factors (Scazzocchio, 2000; Urnov, 2002). Indeed, the light-responsive region (LRR) of light-inducible promoters, e.g., of the al-3 and frq genes, contain GATA or GATA-derived sequences. These regions are recognized in in vitro binding assays by recombinant WC-1 and WC-2 zinc finger domains (Ballario et al., 1996) and by the activated WCC (Froehlich et al., 2002). An in vivo interaction between LRRs and WCC has been recently demonstrated by He and Liu (2005). In some in vitro assays, however, the zinc finger domains of WC-1, NIT-2, and CYS-4 did not preferentially bind to their specific GATA repeats on naked DNA segments consisting of target promoters (Feng and Marzluf, 1998). This suggests that other chromatin components (e.g., histones) may play a role in the regulation of LRR accessibility and may be important in conferring specificity to the response. Nuclear receptors for steroid hormones are, like WC-1, PAS-containing zinc finger proteins with a sensor domain, and they activate transcription only in the presence of specific coactivators, particularly the histone acetyltransferases (HATs) CBP/P300, PCAF, and GCN5 (Hebbar and Archer, 2003). One current model suggests that posttranslational histone modifications constitute a “histone code” that is involved in the control of gene expression and other genetic and epigenetic processes (Turner, 2000; Jenuwein and Allis, 2001).
Evidence for involvement of chromatin modifications in light-inducible transcriptional activation is starting to accumulate (Crosio et al., 2000; Etchegaray et al., 2003; Naruse et al., 2004). Rhythmic acetylation of histone H3 at clock-regulated promoters has been correlated with the circadian activation of clock genes in mammals (Etchegaray et al., 2003; Hastings and Herzog, 2004). A transient burst of histone H3 phosphorylation on residue S10 (H3 S10) has been observed in mammalian hypothalamic suprachiasmatic nuclei after a light pulse (Crosio et al., 2000). In addition, cycles of acetylation and deacetylation of histone H3 and interaction of the transcription factor Clock with the histone acetyltransferase have been demonstrated in mouse (Naruse et al., 2004).
Here, we show evidence for light-dependent transient acetylation of the amino-terminal tail of histone H3 associated with the LRR of the al-3 promoter. The influence of acetylation on light dependent gene expression was confirmed by genetic analyses, both for al-3 and vvd. We also demonstrate that light-inducible H3 acetylation is catalyzed by NGF-1, the homologue of the yeast histone acetyltransferase Gcn5p.
MATERIALS AND METHODS
N. crassa Strains and Culture Conditions
A his-3 strain (matA his-3; FGSC 462) obtained from the Fungal Genetics Stock Center (Kansas City, KS) was used as the recipient strain for transformations to generate strain BG3 (Figure 1). Similarly, a wc-2 strain (allele 234w; FGSC 3817) was used in a control experiment. For chromatin immunoprecipitation (ChIP) experiments, we used N. crassa wild-type strain 74OR23–1A (FGSC 987; Figure 2) as a control. The wc-1 mutant (matA his-3; bd; wc-1null; FGSC 3081) was a gift from J. Dunlap (Dartmouth Medical School, Hanover, NH). Strain N644 (matA; inl am132; [(am/hph/am)ec42pj112]RIP77; Irelan and Selker, 1997) was the transformation host for ectopic insertion of a histone H3 gene bearing a lysine-to-glutamine substitution at residue 14 (hH3K14Q).
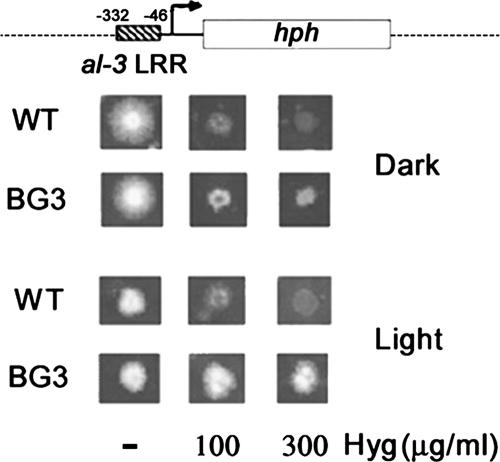
Figure 1.
The al-3 LRR is sufficient to confer light inducibility on the hph reporter gene. Top, schematic representation of the reporter region of pBG3 plasmid, containing the LRR of the al-3 promoter fused to hph, a gene that confers resistance to hygromycin B. The sequence coordinates are relative to the transcription start site. Bottom, host wild-type strain (WT; FGSC462) and a representative transformant (BG3) obtained by transformation with plasmid pBG3 were grown on solid medium plus increasing amounts of Hyg in the dark or light (see Materials and Methods). Only the BG3 strain is able to grow in the presence of Hyg in the light.
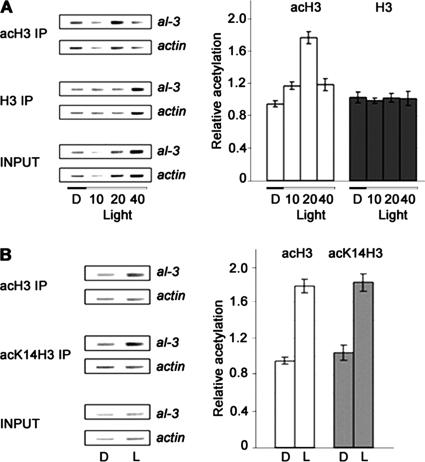
Figure 2.
Light induces a transient increase in acetylation of histone H3. (A) Acetylated histones accumulate 20 min after a light pulse. Left, representative PCR coamplifications of the al-3 LRR and the actin promoter (internal control) from chromatin immunoprecipitations of Neurospora WT (FGSC987) with antibodies directed against acH3 IP or H3 IP. INPUT represents the sample before immunoprecipitation. Right, histograms derived from three independent amplifications with two independent immunoprecipitations. (B) H3 K14 is the target for transient acetylation after a light pulse. Left, representative PCR coamplifications of the al-3 and actin promoter regions 20 min after a light pulse (see A) after immunoprecipitation with antibodies directed against acH3 IP or H3 acetylated specifically on K14 (acK14H3 IP) Right, histograms derived from three independent amplifications with two independent immunoprecipitations.
For dark/light ChIP and Northern experiments, conidia were inoculated in liquid medium and grown in the dark for 18–24 h at 28°C. For dark-grown samples, mycelia were collected by filtration under a red safety lamp and frozen in liquid nitrogen. For illuminated samples, mycelia were photoinduced for 3 min with saturating light (10 W/cm2) and further grown in the dark for different periods. The time intervals indicated in figures are from the beginning of the light pulse to when the cultures were harvested.
Construction of the hH3K14Q Mutant
The hH3K14Q substitution (AAG replaced by CAG at position 14) was generated by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) on pSH12, a plasmid containing the wild-type hH3 gene (Hays and Selker, unpublished data). N644 was cotransformed with pEB11, carrying the hH3K14Q allele and pBT6 (Orbach et al., 1986), a plasmid containing a β-tubulin allele that confers resistance to benomyl (Bml). Transformants were grown en masse in Erlenmeyer flasks on solidified Vogel's sucrose medium containing benomyl. Conidia were plated on media containing no drug, benomyl, or hygromycin (Hyg) to test for reactivation of the hph gene as described previously (Tamaru and Selker, 2001). Random Hyg+ Bml+ transformants were grown in liquid medium to isolate DNA. The presence of ectopic hH3 copies was verified by Southern hybridization. One strain, N3095, was used for further studies.
Repeat-induced Point (RIP) Mutagenesis of ngf-1
We amplified most of the conserved HAT domain of Neurospora GCN Five (ngf-1) with degenerate primers 499 (5′-CTCCCCAAGATGCCCAARGARTAYAT-3′) and 500 (5′-ATRAARTTYTTYGTCCCGAAGTGGTTCCTC-3′) or 501 (5′-CGGCACCCACCTCATGAAYMANYT-3′) and 502 (5′-GGNCTRATRATRCTGCAG-TAGTCCCTGGGGT-3′) and used this as a hybridization probe to identify corresponding cosmid clones in the Orbach/Sachs cosmid library (http://www.fgsc.net/craslib.html#mocosx). Clone G5:H10 contained most of the predicted ngf-1 gene (Supplemental Figure 1). To generate mutants in the ngf-1 gene, we inserted the predicted ngf-1 coding region into pBM60 (Margolin et al., 1997) to yield pBM60-ngf-1. This plasmid was used to insert a second copy of the ngf-1 gene at the his-3 locus by gene replacement (Margolin et al., 1997). Transformants were crossed to induce the premeiotic mutagenic process RIP (Selker, 1990). Progeny were screened for tell-tale restriction fragment length polymorphisms (RFLPs) and DNA methylation by Southern analyses (our unpublished data). One mutant (ngf-1RIP1; N2842) exhibited many RFLPs, and the endogenous copy of the ngf-1 gene was sequenced to identify mutations (Figure 5 and Supplemental Figure 1).
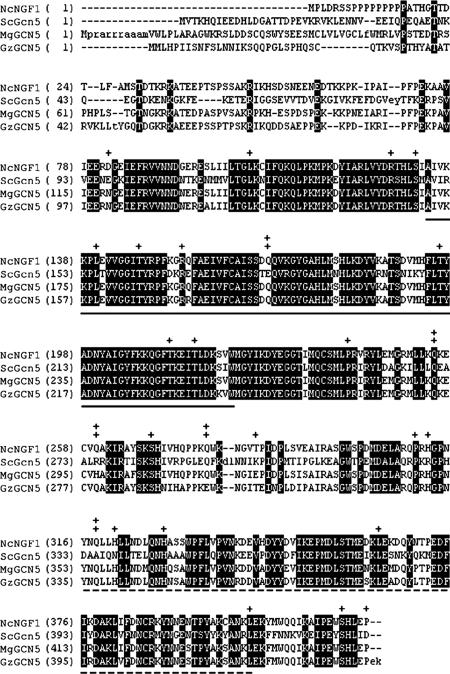
Figure 5.
NGF-1 is the Neurospora homologue of the GCN5 histone acetyltransferase. Alignment of Neurospora NGF-1 (NcNGF1) with homologues from S. cerevisiae (ScGcn5), M. grisea (MgGCN5), and G. zeae (GzGCN5). Bold letters indicate identity among proteins. Mutations resulting in substitutions between wild-type NGF-1 and the predicted amino acid sequence of the mutant allele, ngf-1RIP1, are indicated (+). Predicted nonsense codons are also shown (‡). Solid and dashed underlines indicate the catalytic HAT domain and bromodomain, respectively.
Construction of the Light-responsive al-3-hph Reporter Gene
The promoter region of the al-3 gene (−332 to −46 from the al-3 translational start site) was obtained as a BglII-NruI fragment by polymerase chain reaction (PCR) from the pAL3-ΔNruI plasmid (Carattoli et al., 1991), with primers 5′-GCCCACAGATAGATCTCTTGGCCTTG-3′ and 5′-CCGTCGCGATTATTGGAAACCCGTCGGTA-3′ (the underlined NruI site was added as a primer tail). The product was inserted upstream of the Hyg resistance gene (hph) into BglII + NruI-digested pCH102 carrying a truncated his-3 gene. This plasmid, pBG3, was used to transform strain FGSC462 by electroporation (Margolin et al., 1997). His+ colonies were screened for light-dependent Hyg resistance by spotting equal numbers of conidia of each transformant on minimal medium with or without 100 or 300 μg/ml Hyg. Duplicate plates were kept for 2 d at 28°C in the dark or in a rhythmic regime of 10 min of light (saturating light, 10 W/cm2) and 1 h of dark, to avoid light adaptation.
ChIP Assays
ChIP assays were performed according to a yeast protocol (Avendano et al., 2005) with some modifications. Conidia (106) were used as inocula, and cultures were incubated in liquid medium for 2 d in the dark at 28°C. Dark- and light-induced mycelia were collected by filtration under a red safety light and in vivo cross-linked (Avendano et al., 2005). Cross-linked chromatin–protein complexes were immunoprecipitated with antibodies against unmodified histone H3 (06–755; Upstate Biotechnology, Charlottesville, VA), acetylated H3 (06–599; Upstate Biotechnology), or acetylated H3 K14 (07–353, Upstate Biotechnology). The recovered DNA was subjected to PCR by using the following primers: 30 (5′-AGATAGATCTCTTGGCCTTG-3′) and 31 (5′-CGATTATTGGAAACCCGTCGGTA-3′) for the promoter region of al-3, and ACT1 (5′-CCTCTCTCAGCCAAAGCATC-3′) and ACT2 (5′-GAAAGCTTACCCCATTGTCG-3′) as the internal standard. PCR products were amplified for 25 cycles and resolved on 3% agarose gels. To ensure that the amplified PCR products were in the linear range, the PCR conditions were calibrated with different amounts of immunoprecipitated samples and input DNA (cross-linked chromatin without immunoprecipitation). Band intensities were quantified by optical density analysis with OptyQuant Software (PerkinElmer Life and Analytical Sciences, Boston, MA). As negative controls, mock precipitations were performed in the absence of antibody. PCR products from these negative control samples were not detectable by ethidium bromide staining. The histograms in Figures 2, 3, and 6 represent the ratios between the values for the al-3 and actin PCR products immunoprecipitated, divided by the same ratio obtained by PCR with input DNA [(al-3/act)IP/(al-3/act)input]. Three ChIP replicates were performed on different preparations for each experiment.
Figure 3.
Light-inducible acetylation of H3 K14 requires the presence of WC-1, the blue light photoreceptor. Left, representative PCR coamplifications of al-3 and actin promoter regions (see Figure 2A) after chromatin immunoprecipitation from a wc-1 mutant strain (FGSC3081) with antibodies directed against histone H3 K14 (acK14H3 IP). Right, histograms derived from three independent amplifications with two independent immunoprecipitations.
Figure 6.
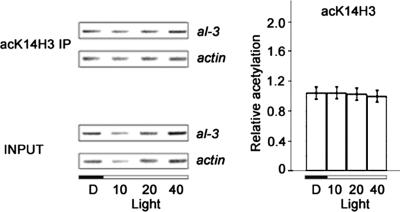
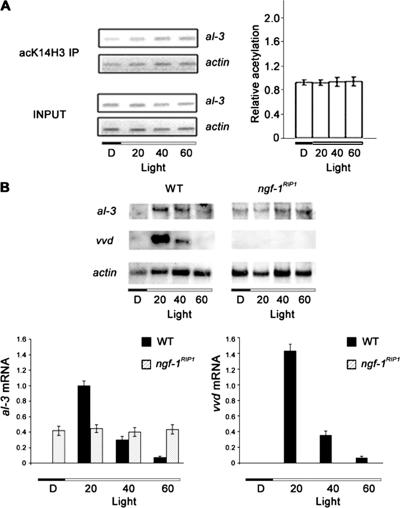
The ngf-1RIP1 mutant is defective in light inducibility. (A) Loss of light-inducible H3 K14 acetylation in the ngf-1RIP1 mutant. Left, representative PCR coamplifications of al-3 and actin promoter regions after immunoprecipitation of chromatin from the ngf-1RIP1 mutant with antibodies directed against histone H3 K14 (acK14H3 IP). Right, histograms derived from three independent amplifications with two independent immunoprecipitations. (B) Loss of mRNA light inducibility in the ngf-1RIP1 mutant. Northern analyses (blots shown on top; densitometric results on bottom) revealed loss of response to light in the ngf-1RIP1 mutant for al-3 and vvd; vvd expression was absent, whereas al-3 mRNA was constitutively expressed in both dark and light conditions.
Northern Assays
Growth conditions were as described for ChIP experiments. RNA was isolated according to a previously published method (Baima et al., 1991). For Northern analyses, 20 μg of total RNA was fractionated on 1.2% agarose gels in 5% formaldehyde-3-(N-morpholino)propanesulfonic acid buffer, transferred to positively charged nylon membranes (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and hybridized according to the manufacturer's instructions. The al-3 transcript was detected with a PCR fragment amplified with primers AL3F (5′-GTCCCTCCAAGGACCTCTTC-3′) and AL3R (5′-CCAGGAGGCTGTGTGTAGCA-3′). The vvd transcript was detected with a PCR fragment amplified with primers VVDF (5′-CCAAACCCCCAAGTAGAACTG-3′) and VVDR (5′-GTCCAGTTCTCTTTCGGTCT-3′). For normalization, the membranes were stripped in 0.1% SDS at 95°C rehybridized with an actin-1 probe obtained by PCR amplification with primers ACTF (5′-GCCTTCTACGTCTCCACCA-3′) and ACTR (5′-GTCGGAGAGACCAGGGTACA-3′). Band intensities were quantified by optical density analysis with OptyQuant Software. The mRNA expression values shown in the histograms of Figures 4 and 6 were calculated as ratios between al-3 or vvd to actin signals in three independent experiments.
Figure 4.
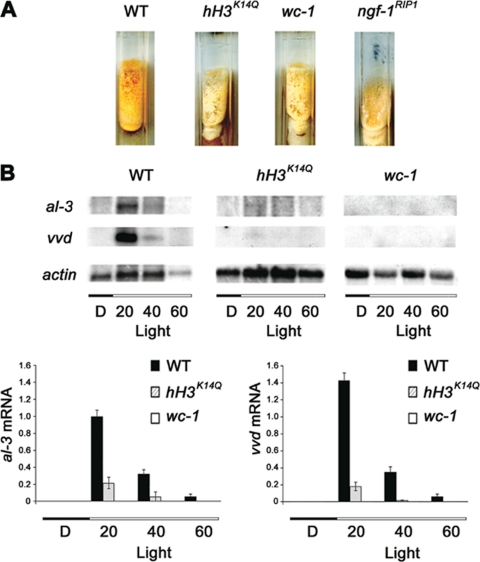
A histone H3 K14Q mutation reduces the light response. (A) Phenotypes of Neurospora strains with wild-type histone H3 (WT; N644) and a dominant histone H3 K14Q substitution (hH3K14Q; N3095). The hH3K14Q mutant phenocopied wc-1 (FGSC3081) and ngf-1RIP1 (N2842). Strains were grown on minimal medium for 1 wk in the dark, induced under saturating light, and the production of carotenoids was observed after 6 h at 4°C. (B) Northern analysis of al-3, vvd, and actin (control) revealed light-induced expression in WT. This regulation was abolished in wc-1 and reduced in hH3K14Q mutants. Densitometric analysis for al-3 (left) and vvd (right) revealed a five- or sevenfold reduction, respectively, in the hH3K14Q strain compared with wild type (below).
Sequence Analysis
Predicted amino acid sequences for NGF-1 and related sequences were compared and aligned with GeneBee Multialignment software (http://www.genebee.msu.su). Sequences used in the alignment were Saccharomyces cerevisiae Gcn5p (AAT93234), Magnaporthe grisea GCN5 (XP_361134), and Gibberella zeae GCN5 (XP_380456). DNA sequences were submitted to the GenBank database (ngf-1, DQ431713; ngf-1RIP1, DQ431714).
RESULTS
The LRR of al-3 Is Sufficient for Light Control of a Reporter Gene
As a first step to investigate the possible involvement of chromatin modifications in light-inducible gene expression in Neurospora, we tested a portion of the al-3 promoter that was predicted to be directly involved in light-dependent transcriptional modifications (Carattoli et al., 1994, 1995). This LRR contains a GATA-X15-GATA motif comparable to binding sites recognized by known GATA factors (Scazzocchio, 2000) and was found to bind recombinant WC-1 or WC-2 zinc-finger domains in an electrophoresis mobility shift assay (Ballario et al., 1996; Linden and Macino, 1997).
We fused the al-3 LRR to the hph reporter gene, which encodes hygromycin phosphotransferase and can result in resistance to Hyg B in Neurospora (Figure 1). Our expression plasmid, pBG3, carries his-3 as a selectable marker for gene targeting. The untransformed recipient wild-type strain grew equally well in the dark and light in the absence of Hyg, but it failed to grow in the presence of Hyg (Figure 1). In contrast, the otherwise isogenic al-3-hph transformant BG3 grew equally well with or without Hyg in the light, showing that the previously defined al-3 LRR is sufficient to confer light inducibility.
Light-induced Acetylation of Histone H3
Previous studies showed that the al-3 gene responds to a light pulse with a transient rapid increase in the level of its mRNA (Baima et al., 1991). Induction is also transient under continuous illumination, but expression decreases more slowly than after a pulse; basal levels are reached after 2 h (Baima et al., 1991; Liu et al., 2003). In our hands, induction of al-3 is maximal at 20 min after the light pulse and transcription returns to basal levels at 60 min. To determine whether a light pulse can influence the degree of acetylation of histone H3 in nucleosomes associated with the LRR, we carried out a series of ChIP experiments (Figure 2). Cross-linked chromatin samples from uninduced (dark; D) and induced (10, 20, or 40 min in light; see Materials and Methods) mycelia were sonicated and used directly (INPUT) or immunoprecipitated with antibodies against histone H3 (H3 IP) or against its acetylated form (acH3 IP). DNA fragments associated with either of the two immunoprecipitations were analyzed by semiquantitative PCR with primers specific for the al-3 LRR and for the promoter of the actin-1 gene, which is not regulated by light. We observed increased acetylated H3 associated with the al-3 LRR 20 min after a light pulse. This acetylation was transient and showed kinetics comparable to that of the al-3 mRNA after induction (Baima et al., 1991). The constant level of histone H3 associated with the LRR suggests that nucleosomes are not depleted from the LRR upon irradiation. The ChIP experiments described above used antibodies directed against H3 peptides acetylated at positions K9 and K14. Acetylation of H3 K14 has been correlated with transcriptional activation in several systems (Carrozza et al., 2003). We therefore considered the possibility that K14 is the target for increased acetylation in the al-3 LRR upon light induction. We compared the levels of al-3 LRR associated with nonspecifically acH3 IP to those precipitated with antibodies directed against H3 acetylated at K14 (acK14H3 IP) in the dark or 20 min after the light pulse (Figure 2B), which corresponds to the time of maximal H3 acetylation found at the al-3 LRR in the first experiment (Figure 2A). We found that the change in H3 acetylation at K14 in the light mirrored that found with K9/K14 acetylated H3. Although we were not able to test K9 separately, our results suggest that H3 K14 is the main, and perhaps exclusive, target of light-inducible histone H3. These findings represent the first evidence that Neurospora light signal transduction involves chromatin modifications.
WC-1, the Blue Light Photoreceptor of Neurospora, Is Required for Light-inducible Acetylation of H3 K14
To determine whether the increase in acetylation at H3 K14 depends on the blue light photoreceptor WC-1, we repeated our ChIP experiments with chromatin isolated from a wc-1 null mutant (Figure 3). In stark contrast to our findings with wild type (Figure 2B, right), we detected no changes in the levels of H3 K14 acetylation at the al-3 LRR in this mutant (Figure 3, right). The same result was obtained with a wc-2 mutant allele (our unpublished data), as expected because an intact WCC is known to be required for appropriate light response in N. crassa (Harding and Turner, 1981). WC-2 was not further investigated in this work, because it does not directly respond to light. This result demonstrates that the H3 K14 modification requires the photoreceptor and establishes an epistatic relationship between WC-1 and the unidentified H3 K14 histone acetyltransferase.
An hH3K14Q Mutant Has wc-like Phenotypes and Exhibits a Loss of Photoinducibility
Our ChIP experiments revealed that a transient light-inducible increase of acetylation at K14 of histone H3 occurs in chromatin associated with the al-3 promoter and that this acetylation depends on the presence of WC-1. The relevance of this chromatin modification was directly evaluated in vivo by construction of a Neurospora strain that carries an ectopic mutant copy of the histone H3 gene (hH3K14Q) engineered to give a lysine to glutamine substitution at residue 14. The lysine was replaced with glutamine because this amino acid is nonacetylable but resembles lysine in size. Additional work with this mutant showed that the mutant histone gene is dominant in another assay (Berge and Selker, unpublished data), and we reasoned that if acetylation of K14 is important for induction by light, this strain might show impaired induction because of having a mixture of wild-type and mutant histone H3, rendering fewer H3 tails available for light-dependent acetylation. Indeed, compared with the isogenic wild type, light-induced carotenogenesis was strongly impaired in the hH3K14Q strain (Figure 4A). This light-induced phenotype resembles that of the wc-1 mutant.
To investigate the possibility that the morphological phenotype was related to transcriptional activation of carotenogenesis genes, we measured al-3 transcript levels in wild-type, wc-1, and the hH3K14Q strains (Figure 4). Induction of al-3 was normal in the strain with the wild-type H3 gene but was absent in the wc-1 mutant, as expected from a previous study (Baima et al., 1992). The hH3K14Q strain showed a fivefold reduction of light inducible al-3 mRNA compared with its parental strain (Figure 4B). Light induction in the hH3K14Q mutant was reduced sevenfold for a second light-inducible gene, vvd (Figure 4B). These results support the suggestion that acetylation of K14 is a requirement for the transcriptional response to light.
The Histone Acetyltransferase NGF-1 Controls Light-induced Acetylation of H3 K14
We wanted to identify the HAT, or HATs, responsible for acetylation of K14 of histone H3 in N. crassa. In the yeast S. cerevisiae, histone H3 is preferentially acetylated by the coactivator Gcn5p (Georgakopoulos and Thireos, 1992). We amplified portions of the Neurospora GCN5 homologue by PCR with degenerate primers based on the conserved HAT domain, screened a Neurospora cosmid library, and sequenced the identified gene, which we called ngf-1. After the N. crassa genome became available, we found this sequence with the program tblastn and S. cerevisiae Gcn5p as bait (Borkovich et al., 2004); its locus number in the most recent genome annotation is NCU10847.2. Overall, the predicted NGF-1 sequence aligns well with known and predicted GCN5 homologues from other fungi (Figure 5). In particular, the catalytic HAT domain (residues 107–219; 78% identity) and the bromodomain (residues 316–401; 63% identity) are conserved between S. cerevisiae Gcn5p and Neurospora NGF-1. Long stretches of C nucleotides are found in the 5′ portion of ngf-1, and this region is not conserved among filamentous fungi (Figure 5), suggesting the existence of an intron. We have attempted, but failed, to isolate complete cDNA. Thus, although most of the gene structure is defined, the DNA sequence upstream of the conserved HAT domain remains uncertain, precluding attempts to generate recombinant NGF-1 protein.
We took advantage of Neurospora's premeiotic mutagenic genome defense system RIP (for review, see Galagan and Selker, 2004) to generate ngf-1 mutants. One mutant, ngf-1RIP1, showed numerous predicted amino acid substitutions and nonsense mutations (Figure 5 and Supplemental Figure 1). The first predicted stop codon (residue 141) should interrupt the catalytic HAT domain. The ngf-1RIP1 mutant grows very poorly, produces few conidia, exhibits colonial growth on sucrose plates, and produces only pale pink mycelia after light induction, all defects that involve the Neurospora light-dependent response. The last phenotype is strikingly similar to that of wc-1 and hH3K14Q mutants (Figure 4A).
We next addressed whether NGF-1 is involved in light-inducible acetylation of H3 K14 (Figure 6A). In contrast to the situation in wild-type strains, no increase of H3 K14 acetylation was observed in the ngf-1RIP1 mutant after light induction (compare Figure 6A with 2). We conclude that NGF-1 is responsible for acetylation of K14 in histone H3 in response to light.
As a separate measure of the involvement of NGF-1 in light regulation, we measured al-3 and vvd transcript levels in the ngf-1RIP1 mutant (Figure 6B). Light inducible expression was lost for both al-3 and vvd, confirming the role of NGF-1 as a coactivator. Unexpectedly, al-3 expression was found to be constitutive in the ngf-1RIP1 strain and its transcript was detected both in the dark and the light, in contrast to the situation in wild type or the semidominant hH3K14Q mutant (compare Figure 6B with 4B). This suggests that the normal light-induced expression of al-3 requires both activation and release of repression.
DISCUSSION
The eukaryotic genome is organized on nucleosomes, the basic units of chromatin. The simple, widely accepted idea that the chromatin constitutes a barrier to the expression of the genes has been refined in the last decade. We now know that nucleosomes can be moved by ATP-dependent remodeling complexes and that histones are substrates for enzymatic modifications (acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, and sumoylation) that can influence gene expression and are in many cases reversible. Histone modifications have been proposed to form an epigenetic code (Strahl and Allis, 2000; Turner, 2000). Modification of histone tails by acetylation of specific lysine residues is regarded as one important mechanism for the regulation of chromatin accessibility to transcription factors and ancillary proteins responsible for global gene expression (Cheung et al., 2000). Here, we report the first evidence for the importance of chromatin acetylation in the well-studied light transduction pathway of Neurospora, a classical organism for photobiology studies. Our findings complement evidence for a relationship between H3 acetylation and rhythmic transcription in a mammalian circadian system (Etchegaray et al., 2003; Curtis et al., 2004). Although a transient burst of histone H3 S10 phosphorylation has been reported in the mammalian hypothalamic suprachiasmatic nuclei after a light pulse (Crosio et al., 2000), we have not observed a statistically significant change in H3 S10 phosphorylation after a light pulse in our system (our unpublished data).
Light-induced Acetylation of K14 of Histone H3
To focus our studies on the critical regions of a light-inducible gene, we first determined that a short region within the photoinducible al-3 promoter, the LRR, is sufficient for light-inducible regulation in vivo (Figure 1). We then discovered that histone H3 associated with this region becomes transiently acetylated after photoinduction (Figure 2A), with kinetics similar to that described for al-3 mRNA induction (Baima et al., 1991). A similar coupling of histone acetylation/transcription induction timing has been also observed for the rhythmic expression of circadian genes (Etchegaray et al., 2003). Lysine 14 of the H3 N-terminal tail is the specific target of the light-dependent modification (Figure 2B), and, as with all other described Neurospora light responses, this modification depends strictly on the presence of the photoreceptor WC-1 (Figure 3). We demonstrated the relevance of the acetylatable H3 K14 residue by genetic manipulation. A strain with a K14Q substitution in histone H3 (hH3K14Q) exhibited a pale pink phenotype strongly resembling that of a wc-1 null mutant (Figure 4A). The hH3K14Q mutant showed a fivefold reduction of induced al-3 mRNA compared with an otherwise isogenic strain bearing only the wild-type hH3 gene (Figure 4B). This marked molecular phenotype was observed in a strain containing both a wild-type and a mutated copy of hH3 gene, indicating that the H3 K14Q mutation is semidominant under these conditions. This result indicates the importance of H3 K14 acetylation in al-3 photoinduction. The requirement of an acetylatable K14 was also demonstrated for another photoinducible gene, vvd (Figure 3B). These findings indicate that chromatin acetylation is a general regulatory step operating in the Neurospora transcriptional photoresponse.
The Neurospora GCN5 homologue NGF-1 Is Responsible for Acetylation of K14 in Response to Light
We wanted to identify the HAT responsible for acetylation of K14 in response to light. Seven putative histone acetyltransferases have been computationally identified in the Neurospora genome (Galagan et al., 2003). Because yeast Gcn5p is known to directly acetylate H3 K14 (Kuo et al., 1996), we tested the possibility that a Neurospora homologoue of this HAT could be responsible for the observed chromatin modification. We isolated the Neurospora GCN5 homologue ngf-1 by homology-based PCR amplification with degenerate primers, sequenced most of the gene, and later also found it in the Neurospora genome sequence, at the terminus of what is now contig 7.2 (http://www.broad.mit.edu/annotation/fungi/neurospora_crassa_7/index.html; Borkovich et al., 2004).
Based on our sequence information, we amplified a large fragment of the ngf-1 gene and used it to generate mutants by RIP (Selker, 1990). Tellingly, the morphological phenotype of the ngf-1RIP1 mutant is similar to the wc phenotype, white mycelium and pale pink conidia (Figure 4A). Moreover, the ngf-1RIP1 showed additional defects, many of which are related to light induction and/or circadian rhythms (e.g., slow growth, reduced number of conidia, and colonial growth) and that may be explained by the general importance of GCN5-type HATs in gene activation (Dyda et al., 2000).
We suggest that NGF-1 is required for light-induced H3 K14 acetylation on the al-3 promoter (Figure 6A). NGF-1 activity depends on the presence of WC-1, because both wc-1 and ngf-1RIP1 mutants are impaired in H3 K14 acetylation (Figures 3 and 6A). Northern analysis of al-3 and vvd mRNA in the ngf-1RIP1 mutant showed loss of photoinduction (Figure 6A), consistent with a role of NGF-1 as coactivator in light-inducible transcription. As expected, we did not observe vvd transcripts either in the dark or upon a light pulse but constitutive al-3 expression occurred in the ngf-1RIP1 mutant. This finding suggests that a NGF-1–mediated repression mechanism may operate on al-3 transcriptional regulation. Together, our results identify NGF-1 as a new element of the Neurospora light transduction system.
Previous studies on transcriptional activation by coactivators such as CBP/p300 have revealed that locus- and time-specific histone modifications are achieved by direct interaction between histone-modifying enzymes and transcriptional regulators (Roth et al., 2001). For example, CBP/p300 interacts with activated nuclear receptors, and this interaction targets HAT activity to specific promoter regions (Tsai and Fondell, 2004). Anafi and colleagues used yeast genetics and in vitro heterologous protein interaction to demonstrate that yeast Gcn5p can regulate a human nuclear receptor (Anafi et al., 2000). In preliminary work to investigate the possibility of direct interactions between WC-1 and a HAT, we carried out a pull-down assay with WC-1 and labeled yeast Gcn5p, reasoning that its high conservation might allow it to mimic its Neurospora homologue that we were unable to express. We found that WC-1 is able to interact in vitro with Gcn5p and that the interaction depends on the region of WC-1 between amino acids 838-1000 (our unpublished data). This region contains a DNA-binding zinc finger domain that has been suggested to be involved in protein–protein interactions (Scazzocchio, 2000). Notably, the WC-1 region required for Gcn5p interaction also contains an AF2 LXXLL motif, which has been shown to be important for the interaction of steroid hormone nuclear receptors with HAT-containing coactivator complexes (Anafi et al., 2000; Savkur and Burris, 2004). The LXXLL motif is also present in a PAS-containing protein that controls the mammalian circadian clock, NPAS2, and its deletion prevents HAT-mediated transcription of clock-regulated genes (Curtis et al., 2004). Our observations are further supported by recent findings from a related system, the time-keeping transcription factor CLOCK is itself a HAT and controls mammalian circadian rhythm by alternative interactions with corepressors and coactivators (Doi et al., 2006).
WCC transiently binds to the promoters of al-3 and other light-inducible genes after illumination (He and Liu, 2005). Notably, this binding follows kinetics similar to what we observed for WC-1-dependent acetylation of H3 at the al-3 promoter. It has also been reported that the form of WCC present in the light (L-WCC) is larger than that observed in the dark (D-WCC) (Froehlich et al., 2002; He and Liu, 2005). L-WCC is thought to owe its increased size to multimerization of WC proteins and/or the presence of unidentified additional factors. In light of our results, we propose that NGF-1 may be recruited in the light by the WCC to form an L-WCC that acts to target the histone acetyltransferase activity on light regulated genes. This could lead to an “open” chromatin structure. Together, our findings reveal a direct link between histone modifications and light transcriptional control and contribute to the developing understanding of the molecular mechanisms operating in light-inducible gene activation.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Corrochiano for the gift of the plasmid pCH102 and J. C. Dunlap for the wc-1 null strain. We thank Z. Lewis for comments. This work was partially supported by Fondazione Pasteur Cenci Bolognetti, Ministero dell'Istruzione, dell'Università e della Ricerca (Fondo per gli Investimenti della Ricerca di Base RBNE01KMT9_009), Progetti Ricerca Interesse Nazionale 2006, and Consiglio Nazionale delle Ricerche RTL (Ricerca a Tema Libero). B.G. was supported by a contract of the University of Rome “La Sapienza.” Work in the laboratory of E.U.S. was supported by U.S. Public Health Service Grant GM-35690 from the National Institutes of Health.
Abbreviations used:
- HAT
histone acetyltransferase
- LRR
light-responsive region
- WCC
White Collar complex.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-03-0232) on August 16, 2006.
REFERENCES
- Anafi M., Yang. Y. F., Barlev N. A., Govindan M. V., Berger S. L., Butt T. R., Walfish P. G. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 2000;14:718–732. doi: 10.1210/mend.14.5.0457. [DOI] [PubMed] [Google Scholar]
- Avendano A., et al. Swi/SNF-GCN5-dependent chromatin remodelling determines induced expression of GDH3, one of the paralogous genes responsible for ammonium assimilation and glutamate biosynthesis in Saccharomyces cerevisiae. Mol. Microbiol. 2005;57:291–305. doi: 10.1111/j.1365-2958.2005.04689.x. [DOI] [PubMed] [Google Scholar]
- Baima S., Carattoli A., Macino G., Morelli G. Photoinduction of albino-3 gene expression in Neurospora crassa conidiation. J. Photochem. Photobiol. 1992;B15:233–238. doi: 10.1016/1011-1344(92)85127-g. [DOI] [PubMed] [Google Scholar]
- Baima S., Macino G., Morelli G. Photoregulation of the albino-3 gene in Neurospora crassa. J. Photochem. Photobiol. 1991;11:107–115. doi: 10.1016/1011-1344(91)80253-e. [DOI] [PubMed] [Google Scholar]
- Ballario P., Vittorioso P., Magrelli A., Talora C., Cabibbo A., Macino G. White collar 1, a central regulator of the blue light responses in Neurospora, is a zinc-finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Cogoni C., Morelli G., Macino G. Molecular characterization of upstream regulatory sequences controlling the photoinduced expression of the albino-3 gene of Neurospora crassa. Mol. Microbiol. 1994;13:787–795. doi: 10.1111/j.1365-2958.1994.tb00471.x. [DOI] [PubMed] [Google Scholar]
- Carattoli A., Kato E., Rodriguez-Franco M., Stuart W. D., Macino G. A chimeric light-regulated amino acid transport system allows the isolation of blue light regulator (blr) mutants of Neurospora crassa. 1995;92:6612–6616. doi: 10.1073/pnas.92.14.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Romano N., Ballario P., Morelli G., Macino G. The Neurospora crassa carotenoid biosynthetic gene (albino 3) reveals highly conserved regions among prenyltransferases. J. Biol. Chem. 1991;266:5854–5859. [PubMed] [Google Scholar]
- Carrozza M. J., Utley R. T., Workman J. L., Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. Light signal transduction in higher plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Cheng P., Yamg Y., Gardner K. H., Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Allis C. D., Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Crosio C., Cermakian N., Allis C. D., Sassone-Corsi P. Light induced chromatin modification in cells of the circadian clock. Nat. Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Curtis A. M., Seo S. B., Westgate E. J., Rudic R. D., Smyth E. M., Chakravarti D., FitzGerald G. A., McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- DeFabo E. C., Harding R. W., Shropshire W. Action spectrum between 260 and 800 nanometers for the photoinduction of carotenoid biosynthesis in Neurospora crassa. Plant Physiol. 1976;57:440–445. doi: 10.1104/pp.57.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M., Hirayama J., Sassone Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;12:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dunlap J. C., Loros J. J. The Neurospora circadian system. J. Biol. Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- Dyda F., Klein D. C., Hickman A. B. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J. P., Lee C., Wade P. A., Reppert S. M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Feng B., Marzluf A. Interaction between major nitrogen regulatory protein NIT2 and pathways-specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa. Mol. Cell. Biol. 1998;18:3983–3990. doi: 10.1128/mcb.18.7.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Fulci V., Macino G. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 2005;56:334–345. doi: 10.1111/j.1365-2958.2005.04545.x. [DOI] [PubMed] [Google Scholar]
- Froehlich A. C., Liu Y., Loros J. J., Dunlap J. C. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Froehlich A. C., Noh B., Vierstra R. D., Loros J. J., Dunlap J. C. Genetic and molecular analysis of Phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell. 2005;4:2140–2152. doi: 10.1128/EC.4.12.2140-2152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Selker E. U. RIP: the evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T., Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding R. W., Turner R. V. Photoregulation of the carotenoid biosynthetic pathway in albino and white collar mutants of Neurospora crassa. Plant Physiol. 1981;68:745–749. doi: 10.1104/pp.68.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M. H., Herzog E. D. Clock genes, oscillators ad cellular networks in the suprachiasmatic nuclei. J. Biol. Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- He Q., Cheng P., Yang Y., Wang L., Gardner K. H., Liu Y. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- He Q., Liu Y. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Shu H., Cheng P., Chen S., Wang L., Liu Y. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J. Biol. Chem. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- Hebbar P. B., Archer T. K. Chromatin remodeling by nuclear receptors. Chromosoma. 2003;111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]
- Huala E., Oeller P. W., Liscum E., Han I. S., Larsen E., Briggs W. R. Arabidopsis NPH 1, a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Irelan J. T., Selker E. U. Cytosine methylation associated with repeat-induced point mutation causes epigenetic gene silencing in Neurospora crassa. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kaldi K., Gonzales B. H., Brunner M. Transcriptional regulation of the Neurospora circadian clock gene wc-1 affects the phase of circadian output. EMBO Rep. 2006;7:119–204. doi: 10.1038/sj.embor.7400595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;19:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Linden H., Macino G. White collar 2, a partner in blue light signal transduction, controlling expression of light regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He Q., Cheng P. Photoreception in Neurospora: a tale of two White Collar proteins. Cell. Mol. Life. Sci. 2003;60:2131–2138. doi: 10.1007/s00018-003-3109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 1997;44:34–36. [Google Scholar]
- Naruse Y., Oh-hashi K., Iijima N., Naruse M., Yoshioka H., Tanaka M. Circadian and light induced transcription of Clock gene Per-1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for α-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol. Cell. Biol. 1986;6:2453–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Denu J. M., Allis C. D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sargent M. L., Briggs W. R. The effects of light on a circadian rhythm of conidiation in Neurospora. Plant Physiol. 1967;42:1504–1510. doi: 10.1104/pp.42.11.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R. S., Burris T. P. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Scazzocchio C. The fungal GATA factors. Curr. Opin. Microbiol. 2000;3:126–131. doi: 10.1016/s1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C., Linden H. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol. Microbiol. 2001;39:1080–1087. doi: 10.1046/j.1365-2958.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Talora C., Franchi L., Linden H., Ballario P., Macino G. Rule of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H., Selker E. U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Turner B. M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Fondell J. D. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam. Horm. 2004;68:93–122. doi: 10.1016/S0083-6729(04)68003-4. [DOI] [PubMed] [Google Scholar]
- Urnov F. D. A feel for the template: zinc finger protein transcription factors and chromatin. Biochem. Cell Biol. 2002;80:321–333. doi: 10.1139/o02-084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.