Abstract
It has recently been reported that the synaptic acetylcholinesterase (AChE) in mosquitoes is encoded by the ace-1 gene, distinct and divergent from the ace-2 gene, which performs this function in Drosophila. This is an unprecedented situation within the Diptera order because both ace genes derive from an old duplication and are present in most insects and arthropods. Nevertheless, Drosophila possesses only the ace-2 gene. Thus, a secondary loss occurred during the evolution of Diptera, implying a vital function switch from one gene (ace-1) to the other (ace-2). We sampled 78 species, representing 50 families (27% of the Dipteran families) spread over all major subdivisions of the Diptera, and looked for ace-1 and ace-2 by systematic PCR screening to determine which taxonomic groups within the Diptera have this gene change. We show that this loss probably extends to all true flies (or Cyclorrhapha), a large monophyletic group of the Diptera. We also show that ace-2 plays a non-detectable role in the synaptic AChE in a lower Diptera species, suggesting that it has non-synaptic functions. A relative molecular evolution rate test showed that the intensity of purifying selection on ace-2 sequences is constant across the Diptera, irrespective of the presence or absence of ace-1, confirming the evolutionary importance of non-synaptic functions for this gene. We discuss the evolutionary scenarios for the takeover of ace-2 and the loss of ace-1, taking into account our limited knowledge of non-synaptic functions of ace genes and some specific adaptations of true flies.
Keywords: acetylcholinesterase, evolution of gene function, Diptera, duplication, gene loss
1. Introduction
The recent discovery of the gene coding of the synaptic acetylcholinesterase (AChE) in mosquitoes has led to a paradox in their evolution. AChE stops neurotransmission in the sensorial synapses of insects by hydrolysing the neurotransmitter acetylcholine (Toutant 1989). The Anopheles gambiae genome contains two ace genes: ace-1, which encodes the main synaptic AChE (Weill et al. 2002) and ace-2, which has an unknown function. These two genes have only 53% similarity at the amino acid level, and the overall ace phylogeny suggests that they diverged before the diversification of the arthropods (Weill et al. 2002). Thus, both genes should be present in most arthropods and have already been formally identified in Hemiptera, Hymenoptera, Lepidoptera and Acari (Weill et al. 2002; Li & Han 2004; Russell et al. 2004; Lee et al. 2006). By contrast, the Drosophila melanogaster genome contains a single gene, ace-2 (Weill et al. 2002), which encodes the synaptic AChE (Fournier et al. 1989). The absence of ace-1 in the D. melanogaster genome can therefore be explained by a secondary loss. Consequently, within the Diptera, either ace-1 (e.g. in mosquitoes) or ace-2 (e.g. in flies) encodes the main AChE, implying that ace-2 took over the function of ace-1 during evolution.
The synaptic AChE is involved in a vital function, and it is assumed that this function has always been present during the evolution of insects. Apparently, AChE has been naturally selected for its particularly high enzymatic activity. It is one of the fastest known enzymes with up to 104 substrate molecules being hydrolysed per second by each enzyme molecule, and its enzymatic velocity seems to be limited only by the diffusion velocity of its substrate (Quinn 1987). This suggests that a slight reduction in its activity would somehow be translated to a significant fitness cost. In the mosquito Culex pipiens, a variant synaptic AChE (coded by ace-1) differing by one amino acid (glycine 119 changed to serine, or G119S) is found in insecticide-treated areas. This variant is insensitive to some insecticides and has a 60% reduced activity, which is associated with substantial fitness cost: about 11% per generation during the breeding season and 50–60% for survival during the overwintering season (Lenormand et al. 1998; Lenormand et al. 1999; Lenormand & Raymond 2000). The G119S mutation has also been detected in insecticide-resistant individuals in distant mosquito species (An. gambiae and An. albimanus, Weill et al. 2004), suggesting that the AChE function cannot be greatly modified (to increase insensitivity to some insecticides) without greatly affecting its optimal and vital activity.
Despite these physiological constraints, the gene encoding the synaptic AChE has apparently changed within the Diptera, with the ancestral gene (ace-1) being replaced by a divergent and distant gene (ace-2). This type of gene replacement, which pertains to a vital function, has never been observed previously (to our knowledge), and thus we have no conceptual framework for understanding how such a phenomenon is possible and what sort of selection has driven it. The Ao (aldehyde oxidase) gene in eukaryotes underwent a possibly related change; it derives neofunctionalization from a duplicate copy of Xdh (xanthine dehydrogenase), and Xdh underwent a second duplication in chordates. The new duplicated copy became a neofunctionalized Ao gene, with the first Ao gene subsequently disappearing from the vertebrate genome (Rodriguez-Trelles et al. 2003). However, the Ao/Xdh and ace-1/ace-2 situations cannot be directly compared because the loss of ace-1 is not associated with a new ace-2 copy, which could have acquired (by neofunctionalization) the same functions as ace-1.
The first step in understanding this situation is to determine which taxonomic groups within the Diptera display this gene change. There are about 129 000 species described within the Diptera order, distributed among 185 families (McAlpine & Wood 1989; Grimaldi & Engel 2005; plus update from M.M.). The main synaptic AChE is encoded by ace-1 in Culex and Anopheles mosquitoes (Culicidae family), and by ace-2 in D. melanogaster. Both the housefly Musca domestica and the olive fruitfly Bactrocera oleae use ace-2 for their cholinergic synapses, as shown by ace-2 mutations providing insecticide resistance (Kozaki et al. 2001a; Walsh et al. 2001; Vontas et al. 2002). Currently, there is no information available on the presence or absence of ace-1 in these two species. Species using ace-2 for their cholinergic AChE belong to distinct families (Drosophilidae, Muscidae, and Tephritidae). This present study aims to determine the presence or absence of both ace-1 and ace-2 in the Diptera families. The information obtained from the study of these genes and their functions allowed us to propose some possible evolutionary scenarios.
2. Material and methods
(a) Insect samples
Diptera species were either collected locally or obtained from various sources (particularly strains and identified preserved materials). Parasite species from the Braulidae, Nycteribiidae and Gasterophilidae families were freshly obtained from host species specialists, respectively (honeybee keeper, chiropterologist and veterinarian). References of samples used are reported in the electronic supplementary material. Most samples were identified by only one person in our team (M.M.).
(b) PCR amplification
DNA extraction was carried out using a DNeasy Tissue Kit (Qiagen) following the manufacturer's instructions. For ace-1, there are published sequences from only one Diptera family (Culicidae), An. gambiae strain KISUMU (AJ515150, exon3; AJ488492, exon 4–9) and strain YAO (AJ515149, exon 3; AJ515148, exon 4–9); Aedes aegypti (AAB35001); C. pipiens strain SLAB (AJ489456) and strain SR (AJ515147). Therefore, we also considered other insect ace-1 sequences: Aphis gossypii strain S171B (AJ748114) and strain S1081K (AJ748115); Schizaphis graminum (Q9BMJ1). All these ace-1 sequences were used to design four pairs of primers: Moustdir (5′ CCG-GGN-GCS-ACY-ATG-TGG-AA 3′) with Moustrev (5′ ACG-ATM-ACG-TTC-TCY-TCC-GA 3′); P6dir (5′ATM-GWG-TTY-GAG-TAC-ACS-GAY-TGG 3′) with P7rev (5′ GGC-AAA-RTT-KGW-CCA-GTA-TCK-CAT 3′); Ace1dir3 (5′ GAC-AAR-ATG-GTS-GGN-GAY-TAT-CA 3′) with Ace1rev4 (5′ CCR-TGC-ATM-ACR-CCN-GTC-CA 3′) or with P7rev. Polymerase chain reaction (PCR) comprised 35 cycles of 93 °C for 30 s, 50 °C for 30 s and 72 °C for 30 s. These four pairs of primers were systematically used for all samples.
We amplified ace-2 using two pairs of degenerate primers: ace2dir4 (5′AAY-GCN-CCS-TGG-AGY-CAY-ATG-AC 3′) with ace2rev6 (5′ CCV-GAR-TAS-GAR-TTC-CAY-TGY-TG 3′), and ace2dir3 (5′ TGG-ATY-TAY-GGB-GGY-GGS-TTY-ATG 3′) with ace2rev3 (5′ GTC-ATR-TGR-CTC-CAS-GGN-GCR-TT 3′). These were designed by comparing published ace-2 sequences from distant Diptera species: D. melanogaster (P07140), M. domestica (Q8MXC4), Lucilia cuprina (P91954), B. oleae (Q8MVZ4), An. gambiae (Q869C3) and C. pipiens (Q86GC8). The first pair of primers was systematically used, with the second being used if the first pair failed.
PCR products were directly sequenced with an ABI prism 310 sequencer using the Big Dye Terminator kit.
(c) Diptera phylogeny
The phylogenetic topology of the major taxonomic divisions in Diptera (infraorders or superfamilies illustrated in figure 1) and the identification of monophyletic groups were established according to published data (table 1; McAlpine & Wood 1989; Wiegmann 1993; Griffiths 1994; Cumming et al. 1995; Oosterbroek & Courtney 1995; Yeates & Wiegmann 1999; Yeates 2002; Grimaldi & Engel 2005).
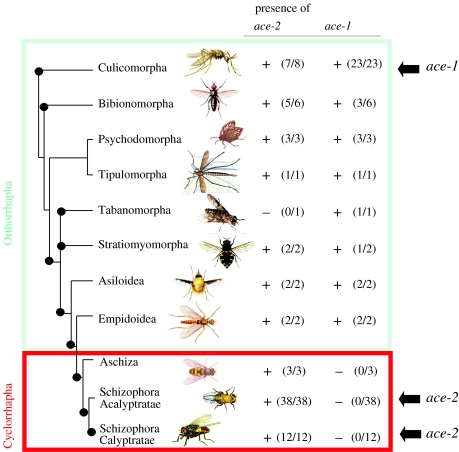
Figure 1.
The distribution of ace genes in Diptera: presence (+) or absence (−) of ace-1 and ace-2 genes in the major divisions of the Diptera phylogeny. Cyclorrhapha or true flies are boxed in red and non-Cyclorrhapha in blue. Ace genes were amplified by PCR and sequenced. The gene is considered ‘absent’ when no amplification occurred after using four (for ace-1) or two (for ace-2) different pairs of primers. ‘Presence’ was always confirmed by sequencing. The number of ‘positive’ species over the total number of species sampled is given in parentheses. For Culicomorpha, data from 15 species investigated in a previous survey (Weill et al. 2002) are also included. Black circles on the phylogeny indicate monophyletic groups. When the information was available, black arrows indicate the ace gene is involved in the synthesis of the main synaptic AChE: ace-1 for five species of Culicomorpha, whereas ace-2 encodes the main synaptic AChE in two species of Schizophora Acalyptratae (Drosophila melanogaster and Bactrocera oleae) and in one species of Schizophora Calyptratae (Musca domestica). See text for explanations.
Table 1.
Taxonomy of the species used for the construction of the Diptera phylogeny using 28S rRNA sequences (fragment D7), and the corresponding accession number. (The outgroup is a flea (order Siphonaptera).)
| infraorder | family | species | accession |
|---|---|---|---|
| Culicomorpha | Culicidae | Culex pipiens | X93403 |
| Chironomidae | Chironomus tentans | X93412 | |
| Simuliidae | Simulium euryadminiculum | X93377 | |
| Bibionomorpha | Bibionidae | Dilophus febrilis | X93375 |
| Psychodomorpha | Psychodidae | Psychoda cinerea | X93404 |
| Scatopsidae | Anapausis inermis | X93374 | |
| Tipulomorpha | Tipulidae | Tipula paludosa | X93405 |
| Stratiomyomorpha | Stratiomyidae | Pachygaster leachii | AF238524 |
| Tabanomorpha | Tabanidae | Tabanus sudeticus | X93371 |
| Asilomorpha | Bombyliidae | Bombylius major | AY456149 |
| Aschiza | Lonchopteridae | Lonchoptera lutea | AF502991 |
| Syrphidae | Rhingia nasica | AF502998 | |
| Schizophora acalyptratae | Otitidae | Ceroxys edwardsii | AF503002 |
| Drosophilidae | Drosophila melanogaster | M21017 | |
| Schizophora calyptratae | Otitidae | Ceroxys edwardsii | AF503002 |
| Muscidae | M. domestica | AJ551427 | |
| Calliphoridae | Chrysomya albiceps | AJ551433 | |
| Calliphoridae | L. cuprina | AJ417709 | |
| Tachinidae | Tachina grossa | AJ300130 | |
| Gasterophilidae | Gasterophilus intestinalis | AJ551429 | |
| (outgroup) | Pulicidae | Archaeopsylla erinace | X93407 |
(d) Test of ace-2 molecular evolution rate
We tested whether there was a faster molecular evolution of ace-2 in groups also possessing ace1 using the following relative rate test procedure. First, 28S rRNA sequences (fragment D7) were downloaded from data banks and used to construct a phylogeny of the Diptera families (electronic supplementary material). The phylogenetic tree was inferred by maximum likelihood (Felsenstein 1981) using a GTR model of nucleotide substitution with a gamma distribution of substitution rates among sites, as implemented in the Phyml program (Guindon & Gascuel 2003). A bootstrap analysis was then carried out using Seqboot (Felsenstein 1993), followed by a Phyml reconstruction. The majority-rule consensus tree was built using PAUP* (Swofford 1998). This topology was then used to connect 27 ace-2 partial sequences included in our sample (indicated in bold, electronic supplementary material), plus 3 ace-2 partial sequences from the literature (Drosophilidae: D. melanogaster, Fournier et al. 1989; Culicidae: C. pipiens and An. gambiae, Weill et al. 2002).
Three outgroups were branched according to the literature (Wheeler et al. 2001; Gaunt & Miles 2002). Likelihood ratio tests were then carried out on amino acids using the PAML program (Yang 1997), according to different evolutionary rate models for the various taxa: a single, global rate for all taxa; three different, local rates for Cyclorrhapha, non-Cyclorrhapha, and outgroups; and one rate for each of the 63 tree branches. Tests were also conducted on nucleotides to detect potential synonymous rate variations among taxa.
(e) Quantification of ace genes expression during development
Larvae of each instar and adults of the C. pipiens SLAB strain were used to extract RNA with Trizol (Life Technologies) and each sample was reverse transcribed. Real time quantitative PCR (Roche light cycler) was used to estimate the number of both ace-1 and ace-2 mRNA copies. Three PCRs were carried out for each developmental stage: one was specific for the ace-1 gene (Moustdir and Moustdrev primers), the second was specific for the ace-2 gene (ace2dir4 and ace2rev6 primers) and the last was specific for the G6PDH gene of which the mRNA expression level remains constant during the different developmental stages of C. pipiens (CpG6PDHdir GCGGCGGGACTTTGAG and CpG6PDHrev AATCCTGTTCCACCCCTTCA primers). Each cDNA template was analysed in triplicate. The ratio between the ace (1 or 2) and G6PDH arbitrary concentrations gave the pattern of expression for both ace genes during the development of C. pipiens.
(f) Obtaining of C. pipiens ace-2 protein in S2 Drosophila cells
5′ and 3′ RACEs were carried out using the ‘GENE RACER’ kit from Invitrogen to give the complete ace-2 cDNA of C. pipiens. The coding cDNA was then amplified using ace2dir ATGTCGTCGATTAGCATGGT and ace2rev GAATAATCTCAGCACGATTA primers and inserted into a pAc5.1/V5-His vector (Invitrogen). S2 cells (20×106) were transfected with the expression vector using Fugene6 (Roche) as a transfection reagent in OptiMEM medium according to the manufacturer's protocol. Cells were maintained in serum free Schneider's medium to avoid endogenous AChE activity due to foetal cow serum. Four days after transfection, the cells were collected and centrifuged at 1200 rpm for 3 min and then homogenized in 500 μl of phosphate buffer (0.25 M) containing 0.1% Triton X100. This was then centrifuged for 10 min at 10 000 rpm and the supernatant used as an AChE2 source.
(g) Detection of ace-1 and ace-2 in the cholinergic activity of C. pipiens
We used two methods to determine the contribution of ace-2 in the cholinergic activity. For method 1, residual activity of AChE from both ace-1 and ace-2 were measured with respect to an increasing concentration of one inhibitor. For method 2, residual activity of AChE from both ace-1 and ace-2 were measured with respect to an increasing time in the presence of an inhibitor. For both methods, fresh samples (larvae, adults or adult heads) were homogenized in a phosphate buffer (0.25 M and pH 7) containing 1% Triton. The homogenates were then centrifuged (12 000g for 5 min) and the supernatants used for detecting enzyme activity. For method 1, extracts from whole larvae, adults and adult heads, and recombinant ace-1 or ace-2 proteins (from C. pipiens, produced in S2 cells) were incubated with 10−4 M of malaoxon (the oxon form of malathion, an organophosphate (OP) insecticide) for various times before adding a substrate solution of acetylthiocholine (10−3 M). For method 2, larvae from two different strains were used, one susceptible (SLAB strain; Georghiou et al. 1966) and one resistant (SR strain, Bourguet et al. 1996) to organophosphates (OP) and carbamate insecticides. Ace-1 recombinant proteins from the SLAB or SR strains were obtained according to Weill et al. (2003). Six inhibitors were tested: propoxur and aldicarb (carbamate insecticides), malaoxon, paraoxon (the oxon form of parathion, OP), trichlorfon (OP) and eserine (an alkaloid from Physostigma venenosum). All inhibitors were purchased from Sigma, except propoxur, which was supplied by Bayer (Leverkusen, Germany). Eight dilutions from the initial concentration (1 M: 10−1–10−8) were used for each inhibitor. AChE residual activity was determined (Ellman et al. 1961) using acetylthiocholine (10−3 M) for each dilution, and was expressed as a percentage of initial activity (without inhibitor) against concentration.
3. Results
(a) Distribution pattern of ace-1 and ace-2 in Diptera
We sampled 78 species representing 50 families (27% of the dipteran families) and 10 infraorders (71% of the dipteran infraorders), and amplified the ace-1 and ace-2 genes using a wide range of degenerated primers to determine the distribution pattern of ace-1 and ace-2 among the different Diptera lineages. We found ace-2 in 75 species, distributed in all major dipteran taxonomic divisions (figure 1). We found no amplified ace-2 in three species: Simulium ornatum (Simuliidae, Culicomorpha), Tabanus bromius (Tabanidae, Tabanomorpha) and Mycetophilidae (Bibionomorpha).
By contrast, we found ace-1 in 21 out of 25 species belonging to the non-Cyclorrhapha group (which includes mosquitoes), but in none of the 53 species of the suborder Cyclorrhapha (or ‘true flies’), despite an intensive PCR investigation. As a control, we were able to amplify ace-2 in all the species lacking ace-1. The Cyclorrhapha, in which we were unable to amplify ace-1, includes the Drosophilidae and in particular D. melanogaster, in which ace-1 is known to be absent.
(b) Evolution of ace-2 within the Diptera
Likelihood ratio tests were carried out for 30 dipteran and 3 outgroup ace-2 cDNA (138 nucleotide sites) and protein (45 amino acid sites) sequences, according to different evolutionary rate models for the various taxa. According to the global clock model, a single evolutionary rate is fixed for all taxa. According to the local clock model, three groups (outgroups, non-Cyclorrhapha and Cyclorrhapha) have their own evolutionary rate. According to the relaxed clock model, each branch of the tree has its own evolutionary rate (i.e. a standard maximum-likelihood analysis). The likelihood of each model is shown in table 2.
Table 2.
Likelihood values of three different models of the molecular evolution of the ace-2 fragment. (See text for explanations.)
| log likelihood | ||
|---|---|---|
| model | cDNA (138 sites) | amino acids (45 sites) |
| global clock | −2736.6 | −1180.7 |
| local clock | −2735.8 | −1178.7 |
| relaxed clock | −2674.1 | −1141.5 |
The log-likelihood values of the relaxed and global clock models were significantly different (χ312=62.5×2=125.0, p<10−5 for cDNA, and χ312=39.2×2=78.4, p<10−5 for proteins). This suggests that substitution rate contrasts accumulated, among the taxa compared, during ace-2 evolution. However, the log-likelihood of the local clock model was not significantly different from the global clock model value (χ22=1.6, p=0.45 for cDNA, and χ22=4.0, p=0.13 for protein). This suggests that the ace-2 evolutionary rate was not different between the Cyclorrhapha (in which ace-1 is absent) and the non-Cyclorrhapha (in which ace-1 is present).
(c) Quantification of ace gene expression during development
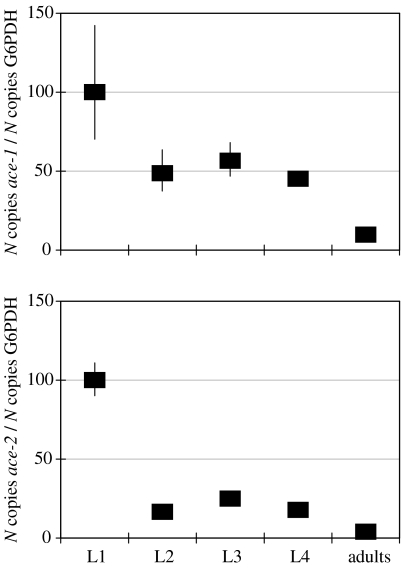
In the mosquito C. pipiens (Culicidae family), ace-2 is expressed, as we were able to quantify mRNA by quantitative PCR in all larval instars (figure 2). From the first instar, ace-1 is also expressed, which is as expected for a gene encoding the main cholinergic AChE. As the levels of ace-1 and ace-2 were determined with different PCR primers, we were unable to directly compare their expression values. Nevertheless, the expression pattern of both genes followed the same trend, with the high level of expression in the newly hatched larvae, a moderately high level from the first instars, and the lowest level of expression in adults.
Figure 2.
Quantitative reverse transcriptase (RT)-PCR carried out at different developmental stages of the mosquito Culex pipiens. The mRNA quantity is expressed as the ratio of the amount of mRNA of the studied gene to the amount of G6PDH mRNA used as a reference, the expression level of which remains constant during development. The value 100 was arbitrarily attributed to the highest ratio value observed. Only the overall profile should be considered when comparing ace-1 and ace-2, see text for explanations.
(d) Detection of ace-2 in the cholinergic activity of C. pipiens
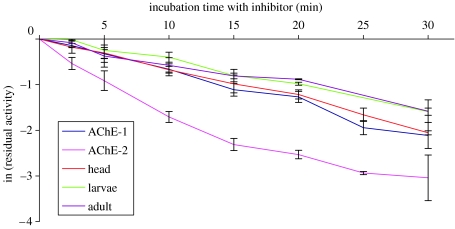
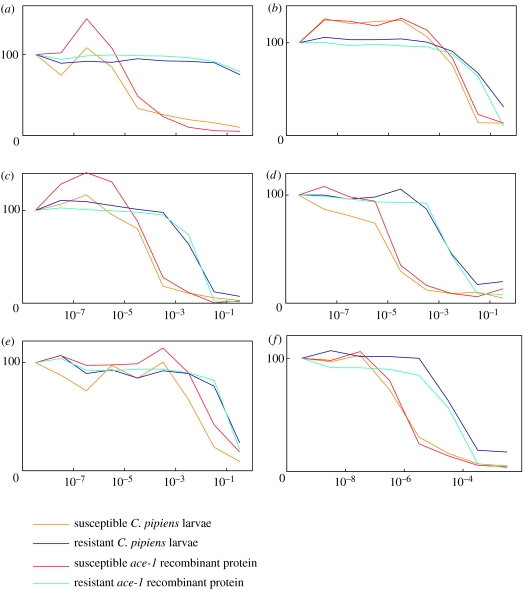
We studied the AChE activity of whole individuals, heads or recombinant proteins from ace-1 and ace-2 in the presence of various AChE inhibitors, which allowed the potential discrimination of enzymatic characteristics, to determine the contribution of ace-2 in the cholinergic AChE activity in a species possessing the two ace genes (C. pipiens mosquito, Culicidae family). The first was used in the same concentration of one inhibitor (malaoxon) with respect to increasing time (method 1). The activity curves obtained from whole larvae, adults and adult heads could be perfectly superimposed over the ace-1 recombinant protein activity curve, and were very different from the ace-2 recombinant protein activity curve (figure 3). This suggested either a very low or a localized enzymatic activity of the ace-2 protein. We then measured the residual activity of AChE from both ace-1 and ace-2 at different concentrations of seven inhibitors (method 2). The activity curves obtained from whole larvae (either the susceptible or the resistant strain) could be superimposed over the corresponding recombinant ace protein activity curve. This suggested that the ace-2 protein was minimally implicated in the main enzymatic activity of mosquitoes. We observed similar results for five inhibitors from two insecticide families (carbamates and OP) and for one plant alkaloid (figure 4).
Figure 3.
Study of AChE residual activity in C. pipiens, expressed as log (A/A0), with A as the activity at time t and A0 as the initial activity, against incubation time (minutes) with malaoxon. Results are shown for recombinant protein AChE1 or AChE2, and for whole larval, adults or head extracts.
Figure 4.
AChE residual activity of C. pipiens larvae and of recombinant ace-1 proteins, (percentage of initial activity), for increasing concentrations of inhibitor (in mol l−1). Both larvae and recombinant proteins are from a susceptible reference strain (SLAB) and an insecticide resistant strain (SR). Six different inhibitors were tested: (a) propoxur, (b) trichlorfon, (c) malaoxon, (d) paraoxon, (e) aldicarb and (f) eserine. See §2 for details. Yellow, susceptible larvae; dark blue, resistant larvae; red, susceptible ace-1 recombinant protein; light blue, resistant ace-1 recombinant protein.
4. Discussion
(a) How many losses of ace-1 within the Diptera?
Diptera divides into paraphyletic lower Diptera (previously Nematocera), paraphyletic lower Brachycera (previously Orthorrhapha) and monophyletic Cyclorrhapha (or true flies). Our data suggest that ace-1 was lost just before the emergence of Cyclorrhapha. This is supported by several points. First, the absence of ace-1 in Cyclorrapha is based on negative PCR amplification, using 4 primer pairs from 53 species, sampled from the three main divisions (Aschiza, Schizophora Acalyptratae and Schizophora Calyptratae), representing 32 out of the 74 families (or 43%) of this taxonomic group. This is consistent with the confirmed absence from whole genome sequence of ace-1 in members of the Drosophilidae family (Schizophora Acalyptratae), i.e. D. melanogaster (Weill et al. 2003) and D. pseudoobscura (P. Fort 2005, personal communication). It is also consistent with M. domestica (Schizophora Calyptratae) and B. oleae (Schizophora Acalyptratae) using ace-2 for the cholinergic AChE. Second, whereas the clade of lower Diptera is paraphyletic, the Cyclorrapha is considered monophyletic, as supported by several unambiguous morphological synapomorphies (McAlpine & Wood 1989; Cumming et al. 1995; Yeates & Wiegmann 1999; Yeates 2002; Grimaldi & Engel 2005) and by molecular data (Wiegmann et al. 2003). Finally, ace-1 is present in one lower Brachycera subdivision that is considered to be closest to the Cyclorrapha (i.e. Empidoidea, electronic supplementary material and figure 1), and is not detected in cyclorraphan families that are close to the lower Brachycera (i.e. Aschiza: Lonchopteridae, electronic supplementary material and figure 1).
For four lower Diptera species, corresponding to the Mycetophilidae, Sciaridae and Stratiomyidae families, ace-1 was not amplified. This could be explained by a large gene divergence that prevents PCR amplification in certain groups. This is strengthened by the successful amplification of ace-1 with another pair of primers in two other species belonging to Mycetophilidae and Stratiomyidae (electronic supplementary material), showing that ace-1 has not been lost in these two families. Thus, the Sciaridea family is the only non-cyclorraphan family (among 18) in which ace-1 was not found, although this family was only represented by one species in our sample. It is possible that outside of the Cyclorrapha, ace-1 has been independently lost in some non-sampled subdivisions (e.g. Ptychopteromorpha, Blephariceromorpha, Axymyiomorpha, Xylophagomorpha, Nemestrinomorpha) or in lower taxonomic units such as the Sciaridae family, although this remains to be established.
In conclusion, ace-1 has been lost in the Cyclorrapha sub order. In this group, ace-2 is probably the only ace gene present, and encodes the cholinergic AChE (as already confirmed in three species). Thus, the replacement of the ace-1 function by ace-2 probably took place around the Jurassic/Cretaceous boundary, as palaeontological evidence suggests that the early cretaceous witnessed the nascent evolution of the Cyclorrapha, with the oldest cyclorraphan fossil being 140 Myr old (Wiegmann et al. 2003).
(b) Understanding of the takeover of ace-2 in Cyclorrapha: proximate considerations
Duplication and subsequent functional divergence of descendant genes has classically been recognized as being the source of new genes (Ohno 1970). Genome analysis has shown that many new functions are associated with gene duplication (He & Zhang 2005). However, the general rules governing functional divergence are unclear. The neofunctionalization hypothesis suggests that after duplication, one daughter gene retains the ancestral function while the other acquires new functions. By contrast, the subfunctionalization hypothesis suggests that the two copies share the ancestral function but differential, tissue-specific expressions. Recent studies have suggested that subfunctionalization is evolutionarily unstable. In other words, it is not generally the terminal fate of the duplicated genes and may evolve towards neofunctionalization, which is evolutionarily stable (He & Zhang 2005; Rastogi & Liberles 2005).
For example, the four ace genes in Caenorhabdis elegans (ace-1, ace-2, ace-3 and ace-4) are the result of three independent duplications. These now code for AChE with different pharmacological and tissue distribution, except for ace-4, in which the new function is still unclear, being either non-catalytic and/or cis-regulating for ace-3 (Combes et al. 2003). In insects, ace-1 and ace-2 (resulting from a duplication occurring before the emergence of the Arthropoda, Weill et al. 2002) have coexisted for a long time. This is probably because they acquired distinct functions (neofunctionalization). So far, the loss of ace-1 occurring at the emergence of Cyclorrhapha is unique because it means that, at least in the cholinergic synapse, ace-2 has retrieved initial functions of the ancestral gene. Thus, it is unclear how to classify this situation according to the various theories of duplication evolution.
The exact function of both ace-1 and ace-2 in non-Cyclorrhapha species must be known in order to propose a valid scenario for gene function replacement. In Culicidae (mosquitoes), which possess both ace genes, AChE activity from ace-2 cannot be detected by enzymatic assays (figures 3 and 4). This suggests either a very low or a very localized enzymatic activity, suggesting that it plays a very small role in synaptic AChE. Thus, according to the literature and the present results, ace-2 only encodes the main synaptic function only in true flies but not in other insects (for a review, see Weill et al. 2002). However, ace-2 is expressed in all larval instars, as we were able to quantify the mRNA by quantitative PCR (figure 2). Also, the sequence conservation of ace-2 across the insects suggests that this gene is being subjected to purifying selection, probably related to restricted synaptic functions and/or other functions. A relative molecular evolution rate test confirmed that the intensity of purifying selection on ace-2 sequences is constant across the Diptera, irrespective of the presence or absence of ace-1 (table 2). Non-synaptic functions have been described for cholinesterases, including developmental involvement in neurogenesis or synaptogenesis in Drosophila (Greenspan et al. 1980; Sternfeld et al. 1998). These non-synaptic functions are probably catalytic, possibly operating through the hydrolysis of the same substrate (acetylcholine or ACh) as the synaptic function (Cousin et al. 2005). This is probably essential for a possible neofunctionalization towards a synaptic function, as it is perhaps the case for ace-2 in true flies. Possible non-synaptic functions of ace-1 are yet to be investigated and if they exist, they may have changed or even disappeared in true flies.
Despite having very little knowledge about the various functions of both ace-1 and ace-2, we know of one requirement for the takeover of ace-2. Both genes must have been co-expressed in cholinergic synapses before the loss of ace-1, and have, or have had, some degree of compensatory function for synaptic ACh hydrolysis. This is because synaptic activity is vitally important and therefore cannot have been interrupted, even temporarily, in the ancestors of true flies. C. elegans presents an example of functional compensation between two ace genes: AChE is supplied at the excitatory neuromuscular junction by both nerve cells, in which one ace gene is expressed, and by muscle cells, in which another ace gene is expressed (Culotti et al. 1981; Johnson et al. 1981; Combes et al. 2003). Here, this functional compensation appears to be stable because it is also found in C. briggsae, which diverged from C. elegans about 30 Myr (ago) (Grauso et al. 1998; Combes et al. 2000). Thus, although functional compensation or co-expression is a necessary condition, it is not sufficient for gene takeover and the subsequent loss of the other gene.
(c) Understanding of the takeover of ace-2 in Cyclorrapha: ultimate considerations
Cyclorrapha is the best defined and most diverse major lineage in Diptera, containing about 105 families and 72 000 species (Grimaldi & Engel 2005; plus update from M.M.). The tremendous success of Cyclorrhapha is linked to several key adaptations, such as fast flying in adults and desiccation resistance of pupae (McAlpine & Wood 1989). There are major differences between Cyclorrapha and other Diptera, particularly within the central nervous system, such as the circumversion (360° rotation) of the male terminalia and a regression of the larval head (McAlpine & Wood 1989; Melzer et al. 1995; Yeates et al. 2002; Grimaldi & Engel 2005). Thus, within the evolution of true flies, there are both innovations and regressions, which offer several possibilities for explaining ace-2 takeover and the loss of ace-1.
A non-synaptic function of ace-1 becoming useless during, for example, an organ regression could trigger the takeover of ace-2, because any non-synaptic function may have provided a strong evolutionary advantage for an ace gene (here, ace-2 in the true flies). This was particularly true when both genes were co-expressed in cholinergic synapses, a possibly unstable situation in which random fluctuation and gene compensation could easily eliminate one of the genes. Ultimately, an ace gene with no non-synaptic function is doomed. There are several possible regression candidates for the loss of a non-synaptic function of ace-1. For example, the regression of the larval eye in true flies and, more generally, the regression of the larval cephalic sensory neurogenesis—the loss of muscle and muscle plaques on the side of the external tergites of the pupa, and of the accompanying nervous system etc. These possible candidates could be evaluated empirically by, for example, determining the level of expression of ace-1 in the tissue of various orthorrhaphan species, particularly families close to the Cyclorrapha.
A phenotypic innovation involving ace-2 could also be proposed. For example, an adaptive change concerning ace-2 (e.g. an increase of AChE activity) could also favour ace-2 in the ace-1/ace-2 activity ratio in synapses. There have been several innovations in true flies that may have triggered a selected change in ace-2. For example, adults have a particularly complex organization of the photoreceptor synapses, characterized by a cholinergic pre-synaptic platform acting as a sort of signal amplification device (Meinertzhagen 1989; Edwards & Palka 1991; Yasuyama & Salvaterra 1999; Buschbeck 2000). This is interpreted as an adaptation to fast flying, which requires particularly efficient and fast receptor–neuron and neuron–neuron communication. In this situation, even a very slight increase in activity could be a significant selective advantage for ace-2, thus changing the ace-1/ace-2 synaptic activity ratio. This could explain the ace-2 takeover of the synaptic function although this requires that ace-1 has no non-synaptic function for it to be subsequently lost.
In conclusion, the loss of ace-1 has occurred in a well-defined taxonomic dipteran group, the Cyclorrhapha. Since they were initially duplicated, ace-1 and ace-2 have coexisted in arthropods and insects for a long time, probably because they acquired distinct functions. So far, the loss of ace-1 at the emergence of Cyclorrhapha is unique because it means that ace-2 must have retrieved, at least in the synapse, the initial functions of the ancestral gene. This event took place about 150 Myr (ago) and therefore cannot be directly investigated. This situation can be further clarified in several ways. In particular, we should identify all of the functions of both genes in non-Cyclorrhapha species to provide useful information that may strengthen or validate one of our possible scenarios involving nervous system innovation or regression in the true flies.
Acknowledgments
We thank N. Agusti, R. Allemand, T. Baldet, D. Bourguet, S. Bréchoire, C. Caceres, A. Callaghan, A. Casteignau, E. Cosson, J.-C. Delécolle, M. Denniff, L. Desprès, T. Disca, G. Duvallet, S. Gouinguene, P. Grébaut, P.-Y. Henry, V. Jamonneau, Y. Layec, B. Mathieu, A. Pomiamkowski, F. Renaud, D. Rey, V. Rufray, F. Schaffner, J. Scott, M. Valero and P. Vernon for Diptera samples, D. Bourguet and N. Pasteur for useful discussion and V. Durand for bibliographic help. This work was financed in part by ANR MoREvol. Contribution 2006.051 of the Institut des Sciences de l'Evolution de Montpellier (UMR CNRS 5554).
Supplementary Material
Table 1. Taxonomy and reference of the samples used. (Accession numbers of the amplified fragment, when obtained, are indicated. Accession numbers in bold were used to perform the test of evolution of ace-2 sequences (see text).)
References
- Bourguet D, Pasteur N, Bisset J, Raymond M. Determination of ace-1 genotypes in single mosquitoes: toward an ecumenical biochemical test. Pest. Biochem. Physiol. 1996;55:122–128. doi: 10.1006/pest.1996.0041. doi:10.1006/pest.1996.0041 [DOI] [PubMed] [Google Scholar]
- Buschbeck E.K. Neurobiological constraints and fly systematics: how different types of neural characters can contribute to a higher level dipteran phylogeny. Evolution. 2000;54:888–898. doi: 10.1111/j.0014-3820.2000.tb00089.x. [DOI] [PubMed] [Google Scholar]
- Combes D, Fedon Y, Grauso M, Toutant J.-P, Arpagaus M. Four genes encode acetylcholinesterase in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. cDNA sequence, genomic structures, mutations and in vivo expression. J. Mol. Biol. 2000;300:727–742. doi: 10.1006/jmbi.2000.3917. doi:10.1006/jmbi.2000.3917 [DOI] [PubMed] [Google Scholar]
- Combes D, Fedon Y, Toutant J.-P, Arpagaus M. Multiple ace genes encoding acetylcholinesterase of Coenorhabditis elegans have distinct tissue expression. Eur. J. Neurosci. 2003;18:497–512. doi: 10.1046/j.1460-9568.2003.02749.x. doi:10.1046/j.1460-9568.2003.02749.x [DOI] [PubMed] [Google Scholar]
- Cousin X, Strähle U, Chatonnet A. Are there non-catalytic functions of acetylcholinesterases? Lessons from mutant animal models. BioEssays. 2005;27:189–200. doi: 10.1002/bies.20153. doi:10.1002/bies.20153 [DOI] [PubMed] [Google Scholar]
- Culotti J.G, von Ehrenstein G, Culotti M.R, Russell R.L. A second class of acetylcholinesterase-deficient mutants of the nematode Caenorhabditis elegans. Genetics. 1981;97:281–305. doi: 10.1093/genetics/97.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming J.M, Sinclair B.J, Wood D.M. Homology and phylogenetic implications of male genitalia in Diptera–Eremoneura. Entomologica Scandinavica. 1995;26:120–151. [Google Scholar]
- Edwards J.S, Palka J. Insect neural evolution-a fugue or an opera? The Neurosci. 1991;3:391–398. [Google Scholar]
- Ellman G.L, Courtney K.D, Andres V, Feather-stone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. doi:10.1016/0006-2952(61)90145-9 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. doi:10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1993 PHYLIP (phylogeny inference package), v. 3.5c. Department of Genetics, University of Washington, Seattle WA.
- Fournier D, Karch F, Bride J.M, Hall L.M.C, Bergé J.-B, Spierer P. Drosophila melanogaster acetylcholinesterase gene, structure, evolution and mutations. J. Mol. Evol. 1989;210:15–22. doi: 10.1016/0022-2836(89)90287-8. [DOI] [PubMed] [Google Scholar]
- Gaunt M.W, Miles M.A. An insect molecular clock dates the origin of the insects and accords with paleontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Georghiou G.P, Metcalf R.L, Gidden F.E. Carbamate-resistance in mosquitoes: selection of Culex pipiens fatigans Wied (=Culex quinquefasciatus) for resistance to Baygon. Bull. World Health Organ. 1966;35:691–708. [PMC free article] [PubMed] [Google Scholar]
- Grauso M, Culetto E, Combes D, Fedon Y, Toutant J.-P, Arpagaus M. Existence of four acetylcholinesterase genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. FEBS Lett. 1998;424:279–284. doi: 10.1016/s0014-5793(98)00191-4. doi:10.1016/S0014-5793(98)00191-4 [DOI] [PubMed] [Google Scholar]
- Greenspan R.J, Finn J.A, Hall J.C. Acetylcholinesterase mutants in Drosophila and their effects on the structure and function of the central nervous system. J. Comp. Neurol. 1980;189:741–774. doi: 10.1002/cne.901890409. doi:10.1002/cne.901890409 [DOI] [PubMed] [Google Scholar]
- Griffiths G.C.D. Relationships among the major subgroups of Brachycera (Diptera): a critical review. Can. Entomologist. 1994;126:861–880. [Google Scholar]
- Grimaldi D, Engel M.S. Cambridge University Press; Cambridge, UK: 2005. Evolution of the insects. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. doi:10.1534/genetics.104.037051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.D, Duckett J.G, Culotti J.G, Herman R.K, Meneely P.M, Russell R.L. An acetylcholinesterase-deficient mutant of the nematode Caenorhabditis elegans. Genetics. 1981;97:261–279. doi: 10.1093/genetics/97.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki T, Shono T, Tomita T, Kono Y. Fenitroxon insensitive acetylcholinesterases of the housefly, Musca domestica, associated with point mutations. Insect Biochem. Mol. Biol. 2001a;31:991–997. doi: 10.1016/s0965-1748(01)00047-9. doi:10.1016/S0965-1748(01)00047-9 [DOI] [PubMed] [Google Scholar]
- Lee D.-W, Kim S.-S, Shin S.W, Kim W.T, Boo K.S. Molecular characterization of two acetylcholinesterase genes from the oriental tobacco budworm, Helicoverpa assulta (Guenée) Biochim. Biophys. Acta. 2006;1760:125–133. doi: 10.1016/j.bbagen.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Lenormand T, Raymond M. Clines with variable selection and variable migration: model and field studies. Am. Nat. 2000;155:70–82. doi: 10.1086/303295. doi:10.1086/303295 [DOI] [PubMed] [Google Scholar]
- Lenormand T, Guillemaud T, Bourguet D, Raymond M. Evaluating gene flow using selected markers: a case study. Genetics. 1998;149:1383–1392. doi: 10.1093/genetics/149.3.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Bourguet D, Guillemaud T, Raymond M. Tracking the evolution of insecticide resistance in the mosquito Culex pipiens. Nature. 1999;400:861–864. doi: 10.1038/23685. doi:10.1038/23685 [DOI] [PubMed] [Google Scholar]
- Li F, Han Z. Mutations in acetylcholinesterase associated with insecticide resistance in the cotton aphid, Aphis gossypii Glover. Insect Biochem. Mol. Biol. 2004;34:397–405. doi: 10.1016/j.ibmb.2004.02.001. doi:10.1016/j.ibmb.2004.02.001 [DOI] [PubMed] [Google Scholar]
- McAlpine J.F, Wood D.M.Manual of nearctic Diptera1989Research Branch Agriculture; Ottawa, Canada [Google Scholar]
- Meinertzhagen I.A. Fly photoreceptor synapses: their development, evolution, and plasticity. J. Neurobiol. 1989;20:276–294. doi: 10.1002/neu.480200503. doi:10.1002/neu.480200503 [DOI] [PubMed] [Google Scholar]
- Melzer R.R, Panzinger A, Reckel F, Smola U. Central nervous system of brachyceran larvae (Insecta, Diptera) Zoologischer Anzeiger. 1995;234:113–123. [Google Scholar]
- Ohno S. Springer; Heidelberg, Germany: 1970. Evolution by gene duplication. [Google Scholar]
- Oosterbroek P, Courtney G. Phylogeny of the Nematocerous families of Diptera (Insecta) Zool. J. Linn. Soc. 1995;115:267–311. doi:10.1006/zjls.1995.0080 [Google Scholar]
- Quinn D.M. Acetylcholinesterase: enzyme structure, reaction dynamics, and virtual transition states. Chem. Rev. 1987;87:955–979. doi:10.1021/cr00081a005 [Google Scholar]
- Rastogi S, Liberles D.A. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 2005;5:28–35. doi: 10.1186/1471-2148-5-28. doi:10.1186/1471-2148-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala F.J. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenasegene. Proc. Natl Acad. Sci. USA. 2003;100:13 413–13 417. doi: 10.1073/pnas.1835646100. doi:10.1073/pnas.1835646100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R.J, Claudianos C, Campbell P.M, Horne I, Sutherland T.D, Oakeshott J.G. Two major classes of target site insensitivity mutations confer resistance to organophosphate and carbamate insecticides. Pest. Biochem. Physiol. 2004;79:84–93. doi:10.1016/j.pestbp.2004.03.002 [Google Scholar]
- Sternfeld M, Ming G.-L, Song H.-J, Sela K, Timberg R, Poo M.-M, Soreq H. Acetylcholinesterase enhances neurite growth and synapse development through alternative contributions of its hydrolytic capacity, core protein, and variable C-termini. J. Neurosci. 1998;18:1240–1249. doi: 10.1523/JNEUROSCI.18-04-01240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. 1998 PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4 Sunderland, MA: Sinauer.
- Toutant J.P. Insect acetylcholinesterase: catalytic properties, tissue distribution and molecular forms. Prog. Neurobiol. 1989;32:423–446. doi: 10.1016/0301-0082(89)90031-2. doi:10.1016/0301-0082(89)90031-2 [DOI] [PubMed] [Google Scholar]
- Vontas J.G, Hejazi M.J, Hawkes N.J, Cosmidis N, Loukas M, Hemingway J. Resistance-associated point mutations of organophosphate acetylcholinesterase, in the olive fruit fly Bactrocera oleae. Insect Mol. Biol. 2002;11:329–336. doi: 10.1046/j.1365-2583.2002.00343.x. doi:10.1046/j.1365-2583.2002.00343.x [DOI] [PubMed] [Google Scholar]
- Walsh S.B, Dolden T.A, Moores G.D, Kristensen M, Lewis T, Devonshire A.L, Williamson M.S. Identification and characterization of mutations in housefly (Musca domestica) acetylcholinesterase involved in insecticide resistance. Biochem. J. 2001;359:175–181. doi: 10.1042/0264-6021:3590175. doi:10.1042/0264-6021:3590175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M, Fort P, Berthomieu A, Dubois M.-P, Pasteur N, Raymond M. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proc. R. Soc. B. 2002;269:2007–2016. doi: 10.1098/rspb.2002.2122. doi:10.1098/rspb.2002.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M, et al. Insecticide resistance in mosquito vectors. Nature. 2003;423:136–137. doi: 10.1038/423136b. doi:10.1038/423136b [DOI] [PubMed] [Google Scholar]
- Weill M, Malcolm M, Chandre F, Mogensen K, Berthomieu A, Marquine M, Raymond M. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 2004;13:1–7. doi: 10.1111/j.1365-2583.2004.00452.x. doi:10.1111/j.1365-2583.2004.00452.x [DOI] [PubMed] [Google Scholar]
- Wheeler W.C, Whiting M, Wheeler Q.D, Carpenter J.M. The phylogeny of the extant hexapod orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. doi:10.1111/j.1096-0031.2001.tb00115.x [DOI] [PubMed] [Google Scholar]
- Wiegmann B.M. Evolutionary origin of the Cyclorrhapha (Diptera): tests of alternative morphological hypotheses. Cladistics. 1993;9:41–81. doi: 10.1111/j.1096-0031.1993.tb00208.x. doi:10.1111/j.1096-0031.1993.tb00208.x [DOI] [PubMed] [Google Scholar]
- Wiegmann B.M, Yeates D.K, Thorne J.L, Kishino H. Time flies, a new molecular time-scale for Brachyceran fly evolution without a clock. Syst. Biol. 2003;52:745–756. doi:10.1080/10635150390250965 [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yasuyama K, Salvaterra P.M. Localization of choline acetyltransferase-expressing neurons in Drosophila nervous system. Microsc. Res. Tech. 1999;45:65–79. doi: 10.1002/(SICI)1097-0029(19990415)45:2<65::AID-JEMT2>3.0.CO;2-0. doi:10.1002/(SICI)1097-0029(19990415)45:2<65::AID-JEMT2>3.0.CO;2-0 [DOI] [PubMed] [Google Scholar]
- Yeates D.K. Relationships of extant lower Brachycera (Diptera): a quantitative synthesis of morphological characters. Zoologica Scripta. 2002;1:105–121. doi:10.1046/j.0300-3256.2001.00077.x [Google Scholar]
- Yeates D.K, Wiegmann B.M. Congruence and controversy: toward a higher-level phylogeny of Diptera. Annu. Rev. Entomol. 1999;44:397–428. doi: 10.1146/annurev.ento.44.1.397. doi:10.1146/annurev.ento.44.1.397 [DOI] [PubMed] [Google Scholar]
- Yeates D.K, David J, Merritt D.J, Baker C.H. The adult ventral nerve cord as a phylogenetic character in brachyceran Diptera. Organ. Divers. Evol. 2002;2:89–96. doi:10.1078/1439-6092-00037 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Taxonomy and reference of the samples used. (Accession numbers of the amplified fragment, when obtained, are indicated. Accession numbers in bold were used to perform the test of evolution of ace-2 sequences (see text).)