Abstract
The flora on the surface of smear-ripened cheeses is composed of numerous species of bacteria and yeasts that contribute to the production of the desired organoleptic properties. Due to the absence of selective media, it is very difficult to quantify cheese surface bacteria, and, consequently, the ecology of the cheese surface microflora has not been extensively investigated. We developed a SYBR green I real-time PCR method to quantify Corynebacterium casei, a major species of smear-ripened cheeses, using primers designed to target the 16S rRNA gene. It was possible to recover C. casei genomic DNA from the cheese matrix with nearly the same yield that C. casei genomic DNA is recovered from cells recovered by centrifugation from liquid cultures. Quantification was linear over a range from 105 to 1010 CFU per g of cheese. The specificity of the assay was demonstrated with DNA from species related to C. casei and from other bacteria and yeasts belonging to the cheese flora. Nine commercial cheeses were analyzed by real-time PCR, and six of them were found to contain more than 105 CFU equivalents of C. casei per g. In two of them, the proportion of C. casei in the total bacterial flora was nearly 40%. The presence of C. casei in these samples was further confirmed by single-strand conformation polymorphism analysis and by a combined approach consisting of plate counting and 16S rRNA gene sequencing. We concluded that SYBR green I real-time PCR may be used as a reliable species-specific method for quantification of bacteria from the surface of cheeses.
Smear-ripened cheeses, such as Munster, Livarot, Maroilles, Limburger, and Tilsit, are characterized by the presence of a complex flora on the surface, comprising many species of yeasts and bacteria. The surface flora has a strong effect on the flavor, texture, and appearance of these cheeses. Yeasts dominate during the early stages of ripening because they are acid tolerant and salt tolerant (7). They increase the pH of the cheese curd by assimilating lactate and producing alkaline compounds, and they also liberate growth factors, thereby favoring the growth of bacterial species. At the end of ripening, the bacteria are dominant, especially the bacteria belonging to the genera Corynebacterium, Brevibacterium, Arthrobacter, and Micrococcus (2, 3, 5, 10, 28). The main sources of surface microorganisms are the milk, the ripening environment, and inoculation of the cheese by use of defined surface cultures or of the so-called “old-young” smearing procedure, in which young cheeses are inoculated with microorganisms from mature cheeses (3).
Problems occasionally occur with the quality of smear cheeses due to the presence of pathogens, such as Listeria monocytogenes (26), and other undesirable microorganisms, such as enterobacteria, enterococci, and molds. A better understanding of the microbial ecology of the cheese surface flora would be very helpful for reducing the occurrence of such problems. It would also be useful for improving control of the beneficial functional properties of the surface flora, such as aroma compound production and color development. However, identification and quantification of cheese surface microorganisms are very difficult. Indeed, many species are present at the same time, and there are almost no selective agar media for any of them. Identification of some bacteria, especially coryneforms, is almost impossible without the use of molecular tools (3).
Real-time PCR is a method based on fluorogenic probes or dyes that is used to determine the copy number of target DNA in a sample. It has been successfully used for quantification of bacteria in various environments (13, 19, 21). However, until now, utilization of this technology for the study or analysis of cheese samples has been very limited.
Rudi et al. (25) were able to detect the presence of viable, dead, or viable but nonculturable cells of L. monocytogenes in Gouda-like cheeses by using a method in which real-time PCR was performed with samples treated with ethidium monoazide bromide. In another study, it was possible to determine the number of copies of the thermonuclease gene of Staphylococcus aureus in artificially contaminated cheeses (14).
The objective of the present study was to demonstrate the usefulness of real-time PCR for quantification of bacteria belonging to the cheese surface flora. To do this, several factors were taken into account. First, a high level of specificity had to be achieved because the cheese surface often contains numerous bacterial species at the same time, several of which are closely related. This is why we chose to develop a method for quantification of Corynebacterium casei, which is a major bacterium of smear cheeses (4) and is frequently present together with other species belonging to the genus Corynebacterium (C. glutamicum, C. ammoniagenes, C. bovis, C. flavescens, and C. variabile). Second, for almost all bacterial species of the cheese surface flora, no or very few DNA sequences are available, except for the gene encoding 16S rRNA, which thus may be considered the most useful gene for designing real-time PCR primers. Third, cheese is a complex matrix from which cells and/or DNA is difficult to isolate, it contains PCR inhibitors (1, 22), and the composition varies depending on the type of cheese and the ripening stage. Consequently, it was essential to validate real-time PCR quantification procedures with representative cheese samples.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and growth conditions used in the present study are described in the supplemental material. Aerobic cultures were grown in 100-ml conical flasks containing 10 ml of medium and inoculated with 2% (vol/vol) from stock cultures stored at −80°C. The flasks were incubated for 48 h on a rotary shaker at 150 rpm. Partial anaerobic cultures were grown in 11-ml tubes containing 10 ml of medium, which were hermetically sealed. They were inoculated and incubated as described above for aerobic cultures, except that they were not agitated.
Extraction of DNA from cheese samples.
Five grams of cheese rind (thickness, 5 mm) was added to a mixture composed of 5 ml of guanidine thiocyanate (4 M) in Tris-HCl (0.1 M, pH 7.5) and 600 μl N-lauryl sarcosine (100 g/liter) and homogenized with a mortar and pestle. A 350-mg aliquot of this mixture was added to a 2-ml tube containing 350 mg of zirconium beads (diameter, 0.1 mm; Sigma, St-Quentin-Fallavier, France), 40 μl of proteinase K (15 mg/ml), and 100 μl of sodium dodecyl sulfate (200 g/liter). The tube was incubated for 2 h in a water bath at 55°C, and 300 μl of sodium phosphate buffer (0.1 M, pH 8), 300 μl of 50 mM acetate-10 mM EDTA buffer (pH 5), and 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1, pH 8) were then added. After cooling on ice, the tube was vigorously shaken in a bead beater (FP120 FastPrep; Savant Instruments Inc., Holbrook, NY) by using three 45-s mixing sequences at a speed of 6 m/s. The tube was cooled on ice for 5 min between mixing sequences. The entire contents of the tube were transferred to a 15-ml tube containing 2.5 ml of a gel in order to improve separation between the liquid and organic phases (Phase Lock Gel Heavy; Eppendorf, Hamburg, Germany) and mixed with 1 ml of phenol-chloroform-isoamyl alcohol. The tube was centrifuged for 3 min at 3,000 × g and 4°C, resulting in separation of the two phases by the gel barrier. Phenol-chloroform-isoamyl alcohol (1.5 ml) was then added to the tube, which was mixed gently, so as not to disturb the gel barrier. After a second centrifugation, 1.5 ml of chloroform was added, and the contents of the tube were mixed and centrifuged a third time. The aqueous phase (approximately 1 ml) was recovered, mixed with 5 μl of RNase A (20 mg/ml; SERVA Electrophoresis GmbH, Heidelberg, Germany), and incubated for 30 min at 37°C. The DNA was then precipitated by adding 100 μl of sodium acetate (3 M, pH 5.2) and 2 ml of ethanol and incubating the tube overnight at −20°C. The DNA was recovered by centrifugation for 15 min at 20,800 × g and 4°C, and the pellet was subsequently washed three times with 2 ml of 70% (vol/vol) ethanol. The pellet was then dried for 15 min in an incubator at 42°C and dissolved in 200 μl of water.
Extraction of DNA from liquid cultures.
Yeast or bacterial cultures (5 ml) were centrifuged for 15 min at 15,000 × g. Each cell pellet was washed with 10 ml of water and resuspended in a mixture composed of 60 μl water, 160 μl guanidine thiocyanate (4 M) in Tris-HCl (0.1 M, pH 7.5), 20 μl N-lauryl sarcosine (100 g/liter), 40 μl proteinase K (15 mg/ml), and 100 μl sodium dodecyl sulfate (200 g/liter). The suspension was then transferred to a 2-ml tube containing 350 mg of zirconium beads. Bead beating and all the subsequent steps were performed as described above for the cheese samples. The DNA concentration was determined with a DU640 spectrophotometer (Beckman, Villepinte, France) at a wavelength of 260 nm. Standard DNA for real-time PCR was obtained by extracting genomic DNA from liquid cultures of C. casei GMPA 2M01.
Production of an external real-time PCR standard by PCR amplification.
The 16S rRNA gene of C. casei was amplified by conventional PCR using primers pA and pH (Table 1). Thermostable Taq DNA polymerase, buffer, and a deoxynucleoside triphosphate mixture were purchased from Takara Biomedicals (Shiga, Japan). The concentrations of primers, Taq DNA polymerase, deoxynucleoside triphosphates, and MgCl2 in the reaction mixture were 0.2 μM, 25 U/ml, 0.2 mM, and 2 mM, respectively. C. casei GMPA 2M01 genomic DNA was added to the PCR mixture at a concentration of 0.1 ng/μl. After an initial denaturation step (5 min at 94°C) and 25 cycles of denaturation (1 min at 94°C), primer annealing (1 min at 57°C), and elongation (2 min at 72°C), the mixture was incubated for 5 min at 72°C. The resulting amplicon was purified by using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations. The absence of nonspecific PCR amplicons was verified by agarose gel electrophoresis and ethidium bromide staining (27), and the DNA concentration was determined at 260 nm. The number of target copies was calculated by assuming that the average molecular mass for 1 bp of double-stranded DNA was 660 g/mol. The calculation was performed by using the following equation: number of copies per microliter = NL × C/mw, where NL is Avogadro's number (6.02 × 1023 molecules per mol), C is the DNA concentration (in g/μl), and mw is the molecular weight of the amplicon (in g/mol) (the size of the amplicon is 1,515 bp).
TABLE 1.
Primers used in this study
| Primer | Target | Application | Sequence (5′-3′) | Reference |
|---|---|---|---|---|
| pA | Eubacteria (16S rRNA gene) | Production of DNA standard and sequencing | AGAGTTTGATCCTGGCTCAG | 9 |
| pH | Eubacteria (16S rRNA gene) | Production of DNA standard and sequencing | AAGGAGGTGATCCAGCCGCA | 9 |
| pB | Eubacteria (16S rRNA gene) | Sequencing | CTCCTACGGGAGGCAGCAGT | 11 |
| pC | Eubacteria (16S rRNA gene) | Sequencing | ACTGCTGCCTCCCGTAGGAG | 11 |
| pG | Eubacteria (16S rRNA gene) | Sequencing | GCATGTGGTTTAATTCGA | 11 |
| pI | Eubacteria (16S rRNA gene) | Sequencing | TCGAATTAAACCACATGC | 11 |
| w34 | Eubacteria (16S rRNA gene) | SSCP | TTACCGCGGCGTGCTGGCAC | 29 |
| w49 | Eubacteria (16S rRNA gene) | SSCP | ACGGTCCAGACTCCTACGGG | 8 |
| fs15 | C. casei (16S rRNA gene) | Real-time PCR | CCGCAAGGCTAAAACTCAAAGGAAT | This study |
| fs17 | C. casei (16S rRNA gene) | Real-time PCR | ACCGACCACAAGGGAAAGACT | This study |
Sequencing of the 16S rRNA gene from selected colony morphotypes.
The 16S rRNA gene of selected morphotypes isolated from commercial smear-ripened cheeses was amplified using primers pA and pH, as previously described. The resulting amplicons were sequenced by Genome Express (Meylan, France), using primers pA and pH and internal primers pB, pC, pG, and pI (Table 1). The sequences were then assembled using the CAP2 program (15) and compared to the GenBank database using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/) to determine the closest known relatives of the 16S rRNA gene sequences.
Alignment of DNA sequences.
16S rRNA gene sequences were aligned using the Pretty program of SeqWeb (version 2) of the Genetics Computer Group (Accelrys, Inc., San Diego, CA). The gene sequences of 121 bacterial species (see the supplemental material) that have been described as species that frequently or occasionally occur in cheeses were considered for primer selection.
Real-time PCR conditions.
SYBR green I PCR amplification was performed using a LightCycler instrument (Roche, Mannheim, Germany). Amplification was carried out in a 20-μl (final volume) mixture containing 5 μl of DNA sample, 4 mM MgCl2, each primer at a concentration of 0.5 μM, and 2 μl of LightCycler-FastStart DNA Master SYBR green I (Roche). All primers were synthesized by Eurogentec (Seraing, Belgium). Amplification involved incubation at 95°C for 8 min for initial denaturation, followed by 45 cycles consisting of (i) denaturation at 95°C for 10 s, (ii) annealing at a temperature that was 5°C below the melting temperature of the primers (except when indicated otherwise below) for 7 s, (iii) extension at 72°C for 9 s, and (iv) fluorescence acquisition (530 nm) at the end of extension. The temperature transition rate was 20°C/s for each step. After real-time PCR, a melting curve analysis was performed by continuously measuring fluorescence during heating from 65 to 95°C at a transition rate of 0.1°C/s. Threshold cycle (CT) values were determined with the LightCycler software (version 3.3), using the second derivative method. Standard curves were generated by plotting the CT values as a function of the log of the initial DNA concentration. PCR efficiency (E) was then calculated using the following formula: E = 10−1/slope (24).
Sequencing of real-time PCR amplicons.
The mixture resulting from real-time PCR amplification was diluted 10,000-fold in water and amplified by conventional PCR as described above, except that amplification was done with primers fs15 and fs17 and primer annealing was done at 65°C for 30 s. The resulting amplicons were sequenced by Genome Express (Meylan, France), using primers fs15 and fs17. The sequences were then assembled as described above.
Addition of C. casei cells to cheese curd.
Pilot-scale production of Livarot-type cheese curd was carried out under aseptic conditions, as described by Leclercq-Perlat et al. (18), and the resulting cheeses were stored at −20°C. After thawing, the cheeses were spiked with C. casei cells. To do this, 5 ml of a culture of C. casei GMPA 2M01 was centrifuged for 15 min at 15,000 × g. The cells were then washed with 10 ml of water and resuspended in 0.3 ml of water. Dilutions of this suspension were prepared and used for addition of 104 to 1010 CFU/g of cheese (the viable cell concentration was determined as described below). DNA was then extracted from cheeses as described above. After real-time PCR analysis, the C. casei concentration was calculated using the following formula: C = A × U × V × R/F, where C is the calculated C. casei concentration in the cheese (in CFU equivalents/g), A is the concentration of C. casei genomic DNA in the DNA sample (prepared from the cheese), determined by real-time PCR (in pg/μl), U is the ratio of the number of CFU subjected to DNA extraction (determined by plate counting) to the final amount of DNA (determined by a spectrophotometric assay at 260 nm) for the C. casei standard DNA prepared from a liquid culture (in CFU/pg), V is the volume of the DNA sample prepared from cheese (in μl), F is the amount of cheese corresponding to volume V (in g), and R is a correction factor taking into account the fact that when DNA is extracted from cheese, the volume of the aqueous phase that can be recovered after phenol-chloroform extraction is slightly less than the volume that can be recovered for the standard DNA prepared from a liquid culture of C. casei. Under our experimental conditions, the U, V, R, and F values were 66.5 CFU/pg, 200 μl, 1.18, and 0.161 g, respectively.
SSCP analyses.
The bacterial community compositions of commercial cheese samples were analyzed by PCR-single-strand conformation polymorphism (SSCP) analyses. Primers w34 and w49 (Table 1) were used to amplify variable region V3 of the bacterial 16S rRNA genes (8). SSCP analyses were performed as previously described (12), except that each PCR amplification was done with 100 ng of DNA sample.
Measurement of viable bacterial concentration.
One gram of cheese rind (thickness, 5 mm) was mixed with 9 ml of physiological saline (9 g/liter NaCl). After dispersion with a mechanical blender (Ultra-Turrax model T25; Ika Labortechnik, Staufen, Germany) for 1 min at 11,500 rpm, 10-fold serial dilutions were prepared in physiological saline and plated on brain heart infusion (BHI) agar supplemented with 50 mg/liter amphotericin (Biokar Diagnostics, Beauvais, France). Colonies were enumerated after incubation for 3 days at 25°C. In contrast to yeasts and molds, most bacteria present on the surface of cheeses are able to form colonies on this medium.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nearly complete 16S rRNA gene sequences of strains 1MA through 1MF and 8MA through 8MF reported in this paper are DQ361012 through DQ361023.
RESULTS
Primer design, calibration curve, and specificity.
To develop a method for quantification of C. casei in which SYBR green I real-time PCR is used, it is necessary to design primers that do not amplify DNA sequences from any other microorganism that may be present in cheese. As is true for most bacterial species isolated from the surface of cheeses, no or very few DNA sequences of C. casei other than the 16S rRNA gene sequence are available. One of the main advantages of using the 16S rRNA gene is that it is possible to identify in silico possible problems of cross-hybridization of the primers designed for C. casei with the DNA of other bacterial species. There is no well-defined procedure for designing specific 16S rRNA gene-targeted primers suitable for SYBR green I real-time PCR assays at this time. For our purposes, the following procedure proved to be convenient. In the first step, the 16S rRNA gene sequences of 121 bacterial species that have been described as species that frequently or occasionally occur in cheeses were aligned as described in Materials and Methods. The C. casei 16S rRNA gene was then examined for the presence of specific regions. For this purpose, we selected all possible regions that were less than 20 nucleotides long and for which there were at least three mismatches with the corresponding regions of all of the 120 other species. Three such regions were found, and they corresponded to positions 71 to 85 (R71-85; 5′ GAATATGACCACTTC 3′), 342 to 356 (R342-356; 5′ CGAAGCCACTTGTGG 3′), and 899 to 916 (R899-916; 5′ GTCTTTCCCTTGTGGTCG 3′) of the C. casei 16S rRNA gene sequence (GenBank accession number AF267152). They were located in variable regions V2, V3, and V6 of the 16S rRNA gene, respectively (23). Real-time PCR primer pairs targeting regions R71-85, R342-356, and R899-916 were then sought using the LightCycler probe design software (v1.0; Roche Applied Science). For each primer pair, only one of the two primers was assigned to target region R71-85, R342-356, or R899-916, whereas the other primer could be freely chosen by the software without any constraints concerning the specificity. When the size of the amplicons was set between 100 and 200 nucleotides and the melting temperature of the primers was set between 62 and 70°C, more than 60 possible primer pairs were found. For each primer pair, the software calculated a score that reflected the presence of motifs such as secondary structures that may have a negative impact on the PCR. The following approach was then used to test primers. First, primer pairs selected from the pairs having the highest scores were used to perform real-time PCR experiments with purified genomic DNA of C. casei and of the other Corynebacterium species previously described as species that occur in cheeses (C. glutamicum, C. ammoniagenes, C. bovis, C. flavescens, and C. variabile). The purified DNA samples were tested at a concentration of 2 ng/μl (10 ng per PCR mixture). The annealing temperature during real-time PCR was set at 5°C below the melting temperature of the primers, as recommended in the LightCycler operator's manual. Only primer pairs for which amplification occurred exclusively with the C. casei DNA were considered to be potentially appropriate for quantitative detection of C. casei. Of the 10 primer pairs that were tested, only 1 was found to fulfill this requirement: primers fs15 and fs17 (Table 1). Primer dimers were often produced with this primer pair, but this was not considered to be a problem since the corresponding threshold cycle was always more than 33. With the other primer pairs, some nonspecific amplification occurred (i.e., there was amplification with DNA from corynebacteria other than C. casei), although there were three mismatches between one of the primers and the corresponding 16S rRNA gene sequence. Increasing the annealing temperature of the real-time PCR did not solve this problem (results not shown). For primers fs15 and fs17, the high level of specificity was probably due to the fact that the mismatches were located near the 3′ end of fs17 (Fig. 1). Furthermore, primer fs17 has only two G or C residues in the last five nucleotides at the 3′ end, which makes it less likely to hybridize transiently and to be available for nonspecific extension by the DNA polymerase (6). The target of primer fs17 includes region R899-916 of the C. casei 16S rRNA gene, which is located within variable region V6.
FIG. 1.
Variable region of 16S rRNA gene used for designing primer fs17. Shading indicates mismatches between fs17 and the 16S rRNA gene sequences of the corynebacteria.
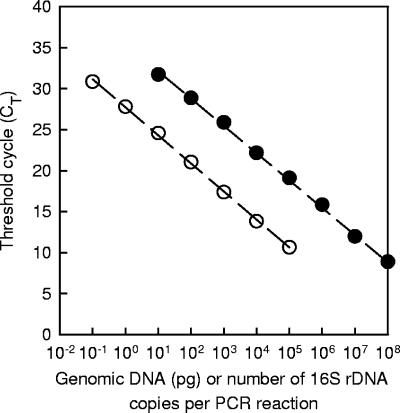
Purified C. casei GMPA 2M01 genomic DNA and the purified PCR product resulting from amplification of the 16S rRNA gene with primers pA and pH were serially diluted 10-fold in water. Five microliters of the dilutions were then subjected to real-time PCR amplification using the reaction and cycling parameters described in Materials and Methods except for the annealing temperature, which was set at 69°C (Fig. 2). This temperature is higher than the melting temperature of primers fs15 and fs17 calculated by the LightCycler probe design software (65°C) and should theoretically have resulted in a low amplification efficiency for the target DNA. However, the amplification efficiency was not lower at 69°C than at 60°C, and thus we used the higher temperature in order to maximize the specificity of the primers. There was a linear relationship between the CT and the logarithm of the copy number (r2 = 0.999 for the data in Fig. 2). The amplification efficiency calculated from these data was 1.98, which is very close to the theoretical maximal yield. With purified genomic DNA, there was also a linear relationship (r2 = 0.999 for the data in Fig. 2), and the PCR efficiency was 1.96. The melting temperature of the C. casei GMPA 2M01 amplicon was 85.5°C. The same temperature was obtained with C. casei DPC 5298. When the amount of genomic DNA and the number of 16S rRNA gene copies were reduced to 0.01 pg and one copy per PCR mixture, respectively, amplifications either did not occur or resulted in primer dimers (with a threshold cycle near 34). In earlier experiments, specific amplification frequently occurred at such low DNA concentrations and also when samples were replaced by water. This was due to slight contamination of the real-time PCR assay preparations with C. casei DNA. Indeed, the sequence of the resulting amplicons was then 100% identical to the corresponding C. casei 16S rRNA gene sequence. This problem was solved by performing DNA extraction, PCR mixture preparation, and post-PCR analysis in separate rooms.
FIG. 2.
Real-time PCR analyses of serial 10-fold dilutions of C. casei genomic DNA (○) or of PCR-amplified 16S rRNA gene (•).
The specificity of the 16S rRNA gene-targeted primers was investigated further with genomic DNA from the bacteria and yeasts listed in the supplemental material (except for the species belonging to the genus Corynebacterium). These organisms are frequently present in cheeses. No amplification occurred (or only primer dimers were produced) in the presence of 104 pg of genomic DNA from these organisms, which further confirmed the specificity of primers fs15 and fs17. Furthermore, in the presence of 104 pg of genomic DNA from C. variabile or Lactococcus lactis, the DNA of C. casei could still be quantified, even at a concentration as low as 1 pg per assay mixture (results not shown). Exogenous DNA therefore did not seem to affect amplification of C. casei DNA with primers fs15 and fs17.
Quantitative detection of C. casei in cheese.
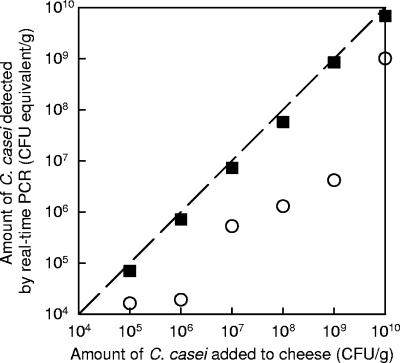
When genomic DNA is extracted from pure cultures of C. casei in BHI broth, as described in Materials and Methods, it is possible to calculate the ratio of the number of CFU subjected to DNA extraction (determined by plate counting) to the final amount of DNA (determined by spectrophotometric assay at 260 nm). We performed five repetitions of extraction on different days and obtained a value of 66.5 CFU per pg of genomic DNA (standard deviation, 2.42 CFU per pg) for the GMPA 2M01 strain. This value was not significantly different from the value obtained with the C. casei DPC 5298 strain (for which the mean value of two repetitions was 70.0 CFU/pg). Theoretically, the concentration of C. casei cells in cheese samples may be calculated from the CT values by using this ratio together with the calibration curve shown in Fig. 2 (see the equation in Materials and Methods). However, such a calculation would be valid only if the DNA recovery yield from C. casei cells in cheese was similar to that from cells recovered by centrifugation from pure cultures in BHI broth. Furthermore, the recovery of DNA should also be independent of the number of C. casei cells present in the cheese sample. This was studied by adding known amounts of C. casei cells to unripened cheese curds, as described in Materials and Methods. Figure 3 shows the amount of C. casei cells detected by real-time PCR (in CFU equivalents per gram of cheese) as a function of the amount added to the cheese. It was observed that quantification was satisfactory only when the DNA samples were diluted at least fivefold before real-time PCR analysis. At higher dilutions of DNA, the detection of C. casei cells was quite similar to that at the fivefold dilution (results not shown). This may be explained by the presence of significant amounts of PCR inhibitors in the undiluted samples. During optimization of the procedure for DNA purification from cheese, we observed that utilization of Phase Lock Gel tubes, as well as thorough washing of the DNA pellets with 70% ethanol three times (as described in Materials and Methods), considerably reduced the impact of PCR inhibitors. However, complete removal of PCR inhibitors was not achieved, which explains the results shown in Fig. 3 for the undiluted samples. Nevertheless, no PCR inhibition was detected for any DNA sample diluted at least five times. This was confirmed in all analyses that we subsequently performed with unripened cheese samples spiked with C. casei cells. Real-time PCR amplifications were also performed after fivefold dilution with DNA samples produced from cheeses spiked with 104 CFU/g of C. casei. In many cases, no amplification occurred. This was probably due to the fact that the amount of target present in the PCR tubes was too small for adequate amplification, as we previously observed with the PCR-amplified 16S rRNA gene standard, when the level was less than 10 copies per PCR mixture. Indeed, with the extraction method described in Materials and Methods, each 1 μl of DNA sample extracted from cheese approximately corresponded to only 0.8 mg of cheese and was further diluted fivefold in order to avoid PCR inhibition. Since reproducible amplification could be obtained with samples spiked with 105 CFU/g of C. casei cells, we estimated that the detection limit of the method was close to 105 CFU/g.
FIG. 3.
Effect of dilution of DNA samples on quantitative detection of C. casei cells in cheese by real-time PCR. Known amounts of bacterial cells were added to fresh cheese curd before DNA extraction. The amount of C. casei cells was calculated from CT values by using the calibration line from Fig. 2 (CT versus amount of genomic DNA) and assuming that 1 pg of genomic DNA corresponds to 66.5 CFU. PCR amplification was performed with nondiluted DNA samples (○) or after fivefold dilution (▪). The dashed line is the equivalence line.
Using Fig. 3, we calculated the percentage of cells detected by real-time PCR in comparison to the number of cells added to the cheese. For the six cheese samples analyzed at a fivefold dilution, the mean ratio was 70% (standard deviation, 8.4%). Thus, we concluded that the level of recovery of DNA from C. casei cells in cheese samples was close to the level of recovery of DNA from pure cultures in BHI broth and was independent of the amount of C. casei cells present in the sample. The DNA extraction and purification procedure described in Materials and Methods limits the influence of the cheese matrix considerably, both for the recovery of DNA and for the effect of PCR inhibitors (based on analysis of fivefold-diluted samples).
To confirm the reproducibility of the quantitative detection of C. casei, the interassay variation was investigated with samples spiked with 2 × 107 C. casei CFU per g of cheese curd. Four separate DNA extractions and real-time PCRs were performed, and the resulting standard error of the C. casei concentration (in CFU equivalents/g) was 16%.
Analyses of commercial smear-ripened cheeses.
The concentrations of bacteria in the rinds of nine French smear-ripened cheeses were determined by plate counting (Table 2). These concentrations were between 2.0 × 108 and 1.0 × 1010 CFU/g. C. casei cells were detected by real-time PCR in six cheeses. The amplicons resulting from real-time PCR amplification with these six cheeses were sequenced as described in Materials and Methods using primers fs15 and fs17. All corresponding sequences were identical to the C. casei sequence, confirming that amplification was specific. Two of the cheeses (cheeses 1 and 8) contained high proportions of C. casei (equivalent to 39 and 42% for cheeses 1 and 8, respectively).
TABLE 2.
Quantification of C. casei in commercial smear-ripened cheeses by real-time PCR
| Sample | Cheese variety | Total bacterial count (CFU/g)a | C. casei concn determined by real-time PCR (CFU equivalents/g)b | C. caseicount/ total bacterial count ratio |
|---|---|---|---|---|
| 1 | Maroilles | 5.4 × 109 | 2.1 × 109 | 3.9 × 10−1 |
| 2 | Munster | 9.3 × 109 | 3.3 × 105 | 3.5 × 10−5 |
| 3 | Munster | 7.5 × 109 | NDc | |
| 4 | Munster | 5.4 × 109 | 1.2 × 106 | 2.2 × 10−4 |
| 5 | Langres | 2.0 × 108 | ND | |
| 6 | Epoisses | 5.3 × 109 | ND | |
| 7 | Livarot | 1.0 × 1010 | 1.5 × 106 | 1.5 × 10−4 |
| 8 | Munster | 7.2 × 109 | 3.0 × 109 | 4.2 × 10−1 |
| 9 | Maroilles | 5.3 × 108 | 3.2 × 106 | 6.0 × 10−3 |
The values are means of three determinations. The standard errors were less than 15%.
The values are means of three determinations, starting with the same DNA sample. The standard errors were less than 15%.
ND, no amplification or production of primer dimers.
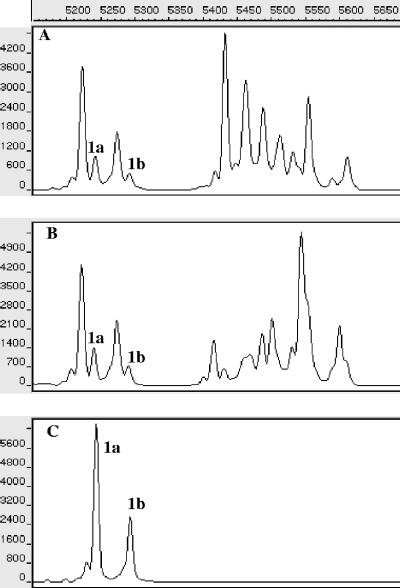
The presence of C. casei cells in cheeses 1 and 8 was confirmed by SSCP analysis of PCR-amplified 16S rRNA gene fragments (Fig. 4). The SSCP pattern obtained for C. casei GMPA 2M01 (Fig. 4C) contained two major peaks (peaks 1a and 1b). These peaks were also present in the SSCP patterns obtained for cheeses 1 and 8, suggesting that the bacterial flora of these cheeses included a high proportion of C. casei. None of the seven other cheeses contained peaks 1a and 1b (results not shown).
FIG. 4.
Region V3 SSCP patterns of PCR-amplified 16S rRNA gene fragments from the bacterial community of cheeses 1 (A) and 8 (B) and from C. casei GMPA 2M01 (C). The y axis indicates fluorescence, and the x axis indicates elution in scans (units of GeneScan software). Peaks 1a and 1b correspond to C. casei.
The presence of C. casei cells in cheeses 1 and 8 was also confirmed by a combined approach consisting of plate counting and 16S rRNA gene sequencing. Six different colony types were distinguished after enumeration of the bacterial flora from cheeses 1 and 8 on BHI agar (Table 3). The nearly complete sequence of the 16S rRNA gene from one representative clone of each morphotype was determined, and the closest relative was determined by comparison to the GenBank database. For each colony morphotype, a partial sequence of the 16S rRNA gene was also determined for three other representative clones, using the sequencing primer pA, in order to monitor the homogeneity within each morphotype. No differences were detected within the morphotypes (results not shown). Two morphotypes from cheese 1 (representative clones 1MA and 1MB) corresponded to the C. casei species (level of sequence identity, 99%). The total concentration of these morphotypes was 3.3 × 109 CFU equivalents/g. This value is not far from the concentration determined by real-time PCR, which was 2.1 × 109 CFU equivalents/g (Table 2). The other colony morphotypes corresponded to Arthrobacter arilaitensis, Marinilactibacillus psychrotolerans, Staphylococcus xylosus, and Brevibacterium linens. For cheese 8, two morphotypes (representative clones 8MA and 8MC) were assigned to the species C. casei and corresponded to a total concentration of 4.2 × 109 CFU/g. This value is not far from the concentration determined by real-time PCR, which was 3.0 × 109 CFU equivalents/g. The four other colony morphotypes were assigned to A. arilaitensis.
TABLE 3.
Identification of the major bacterial flora of two commercial cheeses by 16S rRNA gene sequencinga
| Representative clone | Colony morphotype
|
Colony count (CFU/g) | GenBank accession no. | Closest phylogenic affiliation (GenBank accession no.) | % Identity | |
|---|---|---|---|---|---|---|
| Color | Diam (mm) | |||||
| Cheese 1 | ||||||
| 1MA | Slightly orange | 5 | 2.0 × 109 | DQ361013 | C. casei DPC 5300 (AF267155) | 99 |
| 1MB | Cream | 5 | 1.3 × 109 | DQ361014 | C. casei DPC 5300 (AF267155) | 99 |
| 1MC | Yellow | 5 | 1.2 × 109 | DQ361012 | A. arilaitensis Po102 (AJ609627) | 100 |
| 1MD | Grayish white, semitranslucent | 3 | 8.0 × 108 | DQ361015 | M. psychrotolerans INCT119 (AB159717) | 100 |
| 1ME | White with irregular edge | 8 | 6.7 × 107 | DQ361017 | S. xylosus ATCC 29971 (D83374) | 100 |
| 1MF | Orange | 2 | 3.3 × 107 | DQ361016 | B. linens SB1 (AJ315491) | 99 |
| Cheese 8 | ||||||
| 8MA | Slightly cream | 5 | 3.1 × 109 | DQ361022 | C. casei DPC 5300 (AF267155) | 99 |
| 8MB | Yellow | 5 | 1.3 × 109 | DQ361019 | A. arilaitensisPo102 (AJ609627) | 100 |
| 8MC | Cream | 5 | 1.1 × 109 | DQ361021 | C. casei DPC 5300 (AF267155) | 99 |
| 8MD | Slightly orange | 8 | 1.1 × 109 | DQ361018 | A. arilaitensis Po102 (AJ609627) | 100 |
| 8ME | Greyish white | 3 | 4.0 × 108 | DQ361023 | A. arilaitensisPo102 (AJ609627) | 100 |
| 8MF | Slightly yellow | 5 | 2.3 × 108 | DQ361020 | A. arilaitensis Po102 (AJ609627) | 100 |
For each cheese sample, the different types of colonies grown on BHI agar were enumerated, and the 16S rRNA gene of one representative clone of each morphotype was then sequenced.
DISCUSSION
The present study showed that real-time PCR is a useful method for quantification of bacteria from the cheese surface flora. It is commonly assumed that SYBR green I detection chemistry provides a relative inexpensive and fairly sensitive method for detecting double-stranded DNA (13). Moreover, it does not require specific probes to be developed, as is the case for some other detection chemistries. However, the detection specificity of SYBR green I assays depends entirely on the PCR primers. Using a primer pair targeting the C. casei 16S rRNA gene (primers fs15 and fs17), we were able to ensure a high amplification efficiency for the target sequence and, at the same time, avoid nonspecific amplification of 16S rRNA genes of other species of the cheese surface flora, even species belonging to the genus Corynebacterium. Analysis of the specificity of the 10 different primer pairs that were tested showed that the presence of three mismatches between one of the primers and the corresponding 16S rRNA gene sequence was not always sufficient to avoid nonspecific amplification, even at a high annealing temperature. In order to avoid nonspecific amplification, it was essential that some mismatches were located near the 3′ end of the primers. Moreover, the annealing temperature during assays with primers fs15 and fs17 could be increased to a temperature above the melting temperature calculated by the LightCycler probe design software without decreasing the amplification efficiency. This indicates that the software may sometimes underestimate the melting temperature, and in this case, the specificity of the real-time PCR may have been improved by choosing a higher annealing temperature. The presence of large amounts of exogenous DNA did not affect amplification of the C. casei 16S rRNA gene, which further confirms the high level of specificity of the assay. It is likely that the procedure that we used here to design specific 16S rRNA gene-targeted primers suitable for SYBR green I real-time PCR assays may be used for other bacterial species in the cheese surface flora.
Using the DNA extraction procedure described in Material and Methods, it was possible to obtain yields of genomic DNA from C. casei cells in unripened cheese curd that were nearly the same as the yields from cells recovered by centrifugation from pure cultures in BHI broth. Moreover, the DNA recovery from cheese was linear, at least in the range between 105 and 1010 CFU/g. The cheese matrix thus has a negligible effect on the real-time PCR assay. As a consequence, the real-time PCR assay can be calibrated with serial dilutions of a single DNA sample prepared from C. casei cells grown in BHI broth and recovered by centrifugation. PCR inhibitors were present in DNA samples prepared from cheese, but fivefold dilution was always sufficient to remove PCR inhibition. We recommend that any sample to be analyzed should be assayed using various dilutions of DNA in order to avoid erroneous results due to PCR inhibition. The method for extracting DNA and calibrating the real-time PCR assays can probably be used for other types of cheeses (such as hard cheeses) and bacterial species. This is currently being studied in our laboratory.
In most cases, the bacterial concentration at the surface of smear cheeses after ripening is between 109 and 1010 CFU/g. Since the limit of detection of C. casei cells by real-time PCR is close to 105 CFU/g, it is possible to quantify C. casei as long as it represents between at least 1/10,000 and 1/100,000 of the total bacterial concentration. It is unlikely that the contribution of C. casei to the organoleptic properties of cheeses would be significant at a lower concentration. Nevertheless, it may be possible to improve the sensitivity of the assay. For example, if the DNA purification procedure could be improved so that the amount of PCR inhibitors is reduced, DNA samples could be assayed without fivefold dilution or even after an additional concentration step. In the present study, we had to take several precautionary measures in order to avoid false-positive real-time PCR amplification resulting from contamination of samples or PCR assay mixtures with C. casei DNA. In particular, DNA extraction, PCR mixture preparation, and post-PCR analysis should be performed in separate rooms. Moreover, since cheese samples to be analyzed may be composed of both samples devoid of C. casei and samples containing large amounts of this bacterium, it is important to avoid any cross-contamination of the samples during DNA extraction.
One shortcoming of the quantification method described here is that DNA from dead C. casei cells may also be amplified. Thus, it is not possible to specifically quantify the viable cells. One strategy to overcome this shortcoming could be to perform real-time PCR with samples treated with ethidium monoazide bromide (27).
Using the real-time PCR assay, we detected the presence of C. casei in most of the commercial cheeses that we analyzed, which confirms that this species is a major bacterium on the surface of smear-ripened cheeses (4). However, only two of the nine commercial cheeses contained high concentrations of this bacterium (2.1 × 109 and 3.0 × 109 CFU/g). The presence of C. casei in these samples was confirmed by single-strand conformation polymorphism analysis and by a combination of plate counting and 16S rRNA gene sequencing. The latter approach also revealed the presence of a high concentration of the recently described species A. arilaitensis (16) in the two cheeses. Moreover, one of the cheeses contained 8 × 108 CFU/g of M. psychrotolerans, a marine lactic acid bacterium (17). The presence of this bacterium in smear-ripened cheeses has been reported previously by Maoz et al. (20) and by Feurer et al. (11). M. psychrotolerans is probably well-adapted to the surface of smear-ripened cheeses due to its halophilic and alkaliphilic properties. To our knowledge, strains of C. casei, A. arilaitensis, and M. psychrotolerans are not present in commercial ripening cultures. Their occurrence in cheeses is thus due to contamination from the ripening environment or to the so-called “old-young” smearing procedure.
In summary, this work led to the development of a species-specific method for quantification of C. casei in cheese by SYBR green I real-time PCR. This method should be useful for obtaining a better understanding of the ecology and the functional properties of this bacterium in smear-ripened cheese. A similar quantification procedure can probably be used for most of the other bacterial species found on the surface of smear-ripened cheeses.
Supplementary Material
Acknowledgments
We thank Jean-Pierre Furet and Bertrand Dubreucq for their helpful suggestions.
Footnotes
Published ahead of print on 1 September 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abu Al-Soud, W., and P. Radstrom. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bockelmann, W., and T. Hoppe-Seyler. 2001. The surface flora of bacterial smear-ripened cheeses from cow's and goat's milk. Int. Dairy J. 11:307-314. [Google Scholar]
- 3.Bockelmann, W., K. P. Willems, H. Neve, and K. H. Heller. 2005. Cultures for the ripening of smear cheeses. Int. Dairy J. 15:719-732. [Google Scholar]
- 4.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, N. M., A. C. Ward, T. P. Beresford, P. F. Fox, M. Goodfellow, and T. M. Cogan. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 68:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 7.Corsetti, A., J. Rossi, and M. Gobbetti. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Duthoit, F., J.-J. Godon, and M.-C. Montel. 2003. Bacterial community dynamics during production of registered designation of origin Salers cheese as evaluated by 16S rRNA gene single-strand conformation polymorphism analysis. Appl. Environ. Microbiol. 69:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliskases-Lechner, F., and W. Ginzinger. 1995. The bacterial flora of surface-ripened cheeses with special regard to coryneforms. Lait 75:571-583. [Google Scholar]
- 11.Feurer, C., F. Irlinger, H. E. Spinnler, P. Glaser, and T. Vallaeys. 2004. Assessment of the rind microbial diversity in a farm house-produced vs a pasteurized industrially produced soft red-smear cheese using both cultivation and rDNA-based methods. J. Appl. Microbiol. 97:546-556. [DOI] [PubMed] [Google Scholar]
- 12.Feurer, C., T. Vallaeys, G. Corrieu, and F. Irlinger. 2004. Does smearing inoculum reflect the bacterial composition of the smear at the end of the ripening of a French soft, red-smear cheese? J. Dairy Sci. 87:3189-3197. [DOI] [PubMed] [Google Scholar]
- 13.Hanna, S. E., C. J. Connor, and H. H. Wang. 2005. Real-time polymerase chain reaction for the food microbiologist: technologies, applications, and limitations. J. Food Sci. 70:R49-R53. [Google Scholar]
- 14.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, X. 1996. An improved sequence assembly program. Genomics 33:21-31. [DOI] [PubMed] [Google Scholar]
- 16.Irlinger, F., F. Bimet, J. Delettre, M. Lefevre, and P. A. D. Grimont. 2005. Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int. J. Syst. Evol. Microbiol. 55:457-462. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, M., K. Nakajima, M. Yanagi, Y. Yamamoto, and K. Yamasato. 2003. Marinilactibacillus psychrotolerans gen. nov., sp. nov., a halophilic and alkaliphilic marine lactic acid bacterium isolated from marine organisms in temperate and subtropical areas of Japan. Int. J. Syst. Evol. Microbiol. 53:711-720. [DOI] [PubMed] [Google Scholar]
- 18.Leclercq-Perlat, M. N., A. Oumer, J. L. Bergere, H. E. Spinnler, and G. Corrieu. 1999. Growth of Debaryomyces hansenii on a bacterial surface-ripened soft cheese. J. Dairy Res. 66:271-281. [Google Scholar]
- 19.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 20.Maoz, A., R. Mayr, and S. Scherer. 2003. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 69:4012-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKillip, J. L., and M. Drake. 2004. Real-time nucleic acid-based detection methods for pathogenic bacteria in food. J. Food Prot. 67:823-832. [DOI] [PubMed] [Google Scholar]
- 22.McKillip, J. L., L. A. Jaykus, and M. A. Drake. 2000. A comparison of methods for the detection of Escherichia coli O157:H7 from artificially-contaminated dairy products using PCR. J. Appl. Microbiol. 89:49-55. [DOI] [PubMed] [Google Scholar]
- 23.Neefs, J., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl, M. W. 2004. Quantification strategies in real-time PCR, p. 87-112. In S. A. Bustin (ed.), A-Z of quantitative PCR. International University Line, La Jolla, CA.
- 25.Rudi, K., K. Naterstad, S. M. Dromtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 26.Rudolf, M., and S. Scherer. 2001. High incidence of Listeria monocytogenes in European red smear cheese. Int. J. Food Microbiol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Valdes-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 29.Zumstein, E., R. Moletta, and J. J. Godon. 2000. Examination of two years of community dynamics in an anaerobic bioreactor using fluorescence polymerase chain reaction (PCR) single-strand conformation polymorphism analysis. Environ. Microbiol. 2:69-78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.