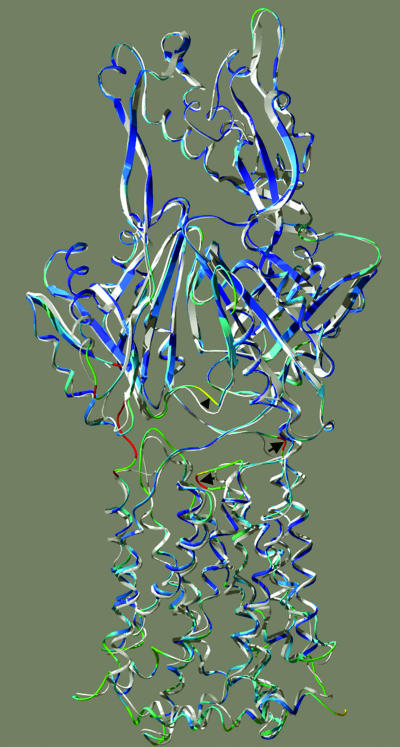
FIG. 5.
Global conformation of AcrB protomers, showing the deviation of the K940A structure from the wild-type structure. The structures were aligned for the best global fit, using backbone atoms, by the Iterative Magic Fit feature of the DeepView program, and the mutant structure was colored in rainbow colors (with red showing the most deviation) by using the “color by RMS deviation” feature of the program, whereas the wild-type structure is shown in white. The structure is shown so that the helices on the right side correspond to the N-terminal ones (TM1 through TM6) and those on the left side correspond to the C-terminal ones (TM7 through TM12). The structure is tilted slightly from the membrane perpendicular in order to show the loop between TM3 and TM4 more clearly. The shift of this loop between TM3 and TM4 is indicated by an arrow on the left, and that of the loop of residues 29 to 34 is shown by an arrow on the right. The shift of the loop at the bottom of the periplasmic ligand binding site (23) is indicated by an arrowhead. Red sections at the extreme left portion of the figure are an artifact caused by the absence of some residues in the wild-type structure. The drawing was done by using DeepView and POV-ray.