Abstract
The presence of the tetracycline resistance determinant tet(M) in human clinical isolates of Escherichia coli is described for the first time in this report. The homologue was >99% identical to the tet(M) genes reported to occur in Lactobacillus plantarum, Neisseria meningitidis, and Streptococcus agalactiae, and 3% of the residues in its deduced amino acid sequence diverge from tet(M) of Staphylococcus aureus. Sequence analysis of the regions immediately flanking the gene revealed that sequences upstream of tet(M) in E. coli have homology to Tn916; however, a complete IS26 insertion element was present immediately upstream of the promoter element. Downstream from the termination codon is an insertion sequence that was homologous to the ISVs1 element reported to occur in a plasmid from Vibrio salmonicida that has been associated with another tetracycline resistance determinant, tet(E). Results of mating experiments demonstrated that the E. coli tet(M) gene was on a mobile element so that resistance to tetracycline and minocycline could be transferred to a susceptible strain by conjugation. Expression of the cloned tet(M) gene, under the control of its own promoter, provided tetracycline and minocycline resistance to the E. coli host.
Tetracyclines are a family of broad-spectrum antibiotics with an excellent safety profile that, some 6 decades after their discovery, still have clinical utility, albeit somewhat limited (12, 32). Tetracyclines have been found to be effective for the treatment of human parasitic diseases and are, in fact, the drug class of choice for treating mefloquine-resistant Plasmodium falciparum infection. Tetracyclines also have a number of nonantibacterial effects that are presumed to play a role in the utility of the drug family for the treatment of periodontal disease and acne (12, 32). The continued spread of tetracycline resistance determinants among clinically important pathogens, aided by nonclinical uses of the compound, has severely limited the clinical utility of the drug class (12, 31, 32).
Tetracycline resistance in bacteria is mediated by four mechanisms: efflux, ribosomal protection, enzymatic inactivation, and target modification (12, 33). Efflux and ribosomal protection are widely distributed among both gram-negative and gram-positive organisms, whereas the other two mechanisms have each been described for only a few bacterial genera (12, 33, 35, 40). Since the first report of transferable tetracycline resistance in Shigella dysenteriae in 1960 (2), 23 genes encoding efflux pumps and 11 genes encoding ribosomal protection proteins have been described for bacteria (12, 31, 33). Ribosomal protection as a mechanism of tetracycline resistance was first reported for streptococci in 1986 (9). The mediators of this resistance mechanism are proteins (e.g., TetM, TetO, TetQ, and TetS) that have the ability to block the binding of and/or displace tetracycline from the 30S subunit of the ribosome (12).
The broad distribution of the ribosomal protection gene tet(M) is due to its association with highly permissive conjugative transposons, such as Tn916 in Streptococcus spp. and Tn1545 in Enterococcus spp. (13). These elements have been identified in over 50 different bacterial genera, both gram negative and gram positive, and have played a major role in the spread of antibiotic resistance determinants between and across genera (12, 13).
Although tet(M) is widely distributed and provides functional resistance to tetracyclines when expressed in Escherichia coli, a tet(M) homologue had not been identified in an environmental or clinical isolate of E. coli until 2004, when it was identified in strains isolated from pigs and chickens (8). The analysis by Bryan et al. (8) examined the presence of 12 tetracycline resistance determinants in natural, nonselected, nonclinical E. coli strains from humans and animal sources and was the first report of the tet(M) determinant in an isolate of E. coli (8).
Tigecycline is the novel 9-t-butyl glycylamido derivative of minocycline that has been approved for clinical use in the treatment of complicated skin and skin structure infections and complicated intra-abdominal infections worldwide (39). During the course of the tigecycline clinical studies, all bacterial isolates were screened for susceptibility to tetracycline and minocycline among a panel of other antibiotics. Isolates of E. coli that were resistant to minocycline and/or tetracycline (MIC ≥ 8 μg/ml) were screened by PCR for the presence of tetracycline resistance determinants, including tet(M). Three isolates from two different patients were found to carry tet(M). This is the first report of the identification of the tet(M) tetracycline resistance determinant originating in a human clinical isolate of E. coli.
MATERIALS AND METHODS
Bacterial strains.
Clinical isolates of E. coli (Table 1) were from the phase 3 double-blind clinical trials comparing the safety and efficacy of tigecycline to those of an active comparator for the treatment of complicated intra-abdominal infections (2002 to 2004) (6). Patient specimens were processed and bacterial pathogens were cultured by each site laboratory according to local practices. Individual investigators sent all bacterial isolates to a central laboratory for identification and susceptibility testing.
TABLE 1.
E. coli strains used in study
| Strain | Characteristic(s) | Source or referencea |
|---|---|---|
| NCTC 50268 | tet(A) | NCTC |
| NCTC 50365 | tet(B) | NCTC |
| NCTC 50270 | tet(C) | NCTC |
| NCTC 50271 | tet(D) | NCTC |
| NCTC 50272 | tet(E) | NCTC |
| GC2270 | tet(M)b | Wyeth Collection |
| DH5α | Cloning strain | 18 |
| GAR3139 | tet(M) tet(A) | 7 |
| GAR3141 | tet(M) tet(A) | 7 |
| GAR3142 | tet(M) tet(A) | 7 |
| GAR7071 | Tetracycline susceptible, levofloxacin resistant | 7 |
| GAR7090 | Tetracycline susceptible, levofloxacin resistant | 7 |
| GC7939 | tet(M), tet(A) transconjugant; GAR3139 donor, GAR7071 recipient | This study |
| GC7940 | tet(M) tet(A) transconjugant; GAR3139 donor, GAR7090 recipient | This study |
| GC7941 | tet(M) tet(A) transconjugant; GAR3141 donor, GAR7071 recipient | This study |
| GC7942 | tet(M) tet(A) transconjugant; GAR3141 donor, GAR7071 recipient | This study |
| GC7949 | tet(M) cloned into pCR-XL-TOPO | This study |
NCTC, National Collection of Type Cultures (www.hpa.org.uk).
S. aureus tet(M) cloned into pUC19 (Wyeth, unpublished data).
Susceptibility determination.
Susceptibility tests were performed by broth microdilution with Mueller-Hinton II broth (MHB) as recommended by the Clinical Laboratory Standards Institute (CLSI [formerly the NCCLS]) (14, 24). A screening test for resistance to tetracycline was also performed using the disk diffusion method according to standard protocols (23).
Amplification of tetracycline resistance determinants.
In order to monitor the presence of resistance determinants for tetracyclines (minocycline and tetracycline), PCR assays were developed and optimized using published sequence information (3, 4, 21, 27, 34) and sequence information directly deposited in GenBank (www.ncbi.nlm.nih.gov) for primer design. Primer sequences, primer location (starting base pair), and expected amplicon size are shown in Table 2. In addition to the resistance determinant, a primer pair specific for 16S rRNA was also included for internal standardization and quality control of the assay (30). Control strains that were previously characterized with respect to antibiotic resistance and the presence of specific determinants were utilized for assay development (Table 1). DNA was obtained from whole-cell lysates as follows. E. coli isolates were plated on Luria agar (Becton Dickenson and Company, Cockeysville, MD), and following overnight incubation, several colonies were collected with a 10-μl loop, resuspended in 500 μl of distilled water, and incubated for 5 min in a boiling water bath. The lysate was subjected to brief centrifugation (Savant SFA13K; Savant, Farmingdale, NY) at 13,000 × g, and 1 μl of the supernatant was used as the template for amplification.
TABLE 2.
Primers used in this study
| Target | Sequence (5′-3′)a | Start point (bp)b | Amplicon size (bp) | Reference sequence accession no.c |
|---|---|---|---|---|
| tet(A) | F: 5′ GTA ATT CTG AGC ACT GTC GC | 25 | 956 | AJ313332 |
| R: 5′ CTG CCT GGA CAA AAT TGC TT | 981 | |||
| tet(B) | F: 5′ GTT ACT CGA TGC CAT GGG GA | 36 | 1,121 | AB089594 |
| R: 5′ GAA GGT CAT CGA TAG CAG GA | 1157 | |||
| tet(C) | F: 5′ GCG CTR TAT GCRDTG ATG C | 148 | 748 | J01749 |
| R: 5′ TGG TCG TCA TCT ACC TGC | 996 | |||
| tet(D) | F: 5′ GCG CTR TAT GCRDTG ATG C | 142 | 748 | X65876 |
| R: 5′ CAT CCG GAA GTG ATA GC | 890 | |||
| tet(E) | F: 5′ GCG CTR TAT GCRDTG ATG C | 162 | 849 | L06940 |
| R: 5′ CTA CCT GAC CGA CAC G | 1011 | |||
| tet(M)d | F: 5′ ATA GAY ACG CCA GGM CAT A | 698 | 1,070 | M21136 |
| R: 5′ GGA GCC CAG AAA GGA TTY GG | 1768 | |||
| ectet(M)up | F: 5′ GTG ATT CTA AAG TAT CC | 342 | 586 | M21136 |
| R: 5′ TAG GAT ACA GTT CTA CC | 928 | |||
| ectet(M)dwn | F: 5′ CAA GAA AAG TAT CAT GTG G | 1666 | 737 | M21136 |
| R: 5′ TTT CAT CTT ATT TAA CAA GAA ACC | 2403 | |||
| ectet(M)up1000 | F: 5′ CAA CTA TCA TAG AAA AGG AAT ACG | 11016 | 1,004 | U09422 |
| R: 5′ TTC CCA CTG AAA AGA GGT TAT TCC | 630 | M21136 | ||
| ectet(M)up500 | F: 5′ TCA AGC TCT ATC CTA CAG C | 11502 | 532 | U09422 |
| R: 5′ TTC CCA CTG AAA AGA GGT TAT TCC | 630 | M21136 | ||
| ectet(M)dwn500 | F: 5′ ATA GTC GGA TAG ATA AAG TAC G | 2353 | 475 | M21136 |
| R: 5′ AAC TTG GTA AAA AGC ACC C | 14495 | U09422 | ||
| ectet(M)dwn1000 | F: 5′ ATA GTC GGA TAG ATA AAG TAC G | 2353 | 955 | M21136 |
| R: 5′ GAC AAG AAC CCA ATG TAA GG | 14973 | U09422 | ||
| ectet(M)clone | F: 5′ TTA CAA ATA TGC TCT TAC GTG C | 152 | 2,251 | M21136 |
| R: 5′ TTT CAT CTT ATT TAA CAA GAA ACC | 2403 | |||
| 16S rRNA | F: 5′ GCCAGCAGCGCGGTAATACG | 537 | 271 | M87484 |
| R: 5′ GGACTACCAGGGTATCTAATCC | 808 |
IUB codes (underlined) are as follows: R, A + G; Y, C + T; D, G + A + T; and M, A + C. F, forward; R, reverse.
Numbering is per the referenced sequence.
GenBank accession numbers (www.ncbi.nlm.nih.gov).
Primers were modified from reference 34.
The FAILSAFE PCR system (Epicenter Technologies, Madison, WI) was used for amplification. Appropriate buffers were experimentally identified for each primer set. One microliter of the whole-cell lysate was used in the PCR assay in a 25-μl volume reaction mixture. Cycling conditions were as follows: the initial denaturation step was for 5 min at 94°C; amplification was 30 cycles of 1 min at 56°C, 1 min at 72°C, 1 min at 94°C, and a final 1 min at 45°C; and extension was for 5 min at 75°C. The reaction products were resolved on a 0.8% agarose gel containing 0.5 μg/ml ethidium bromide. A 1-kb ladder (Fermentas, Hanover, MD) was run on each gel as a size reference.
Sequence determination.
PCR amplicons were TA cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA) for sequence analysis using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit mix (version 3.1; Applied Biosystems, Foster City, CA). Amplification reactions were performed on a model PTC-225 cycler (Bio-Rad, Hercules, CA) for 40 cycles (96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 2 min), and the excess dye was removed by gel filtration on a 96-well Performa DTR plate (from EdgeBiosystems, Gaithersburg, MD). The samples were heat denatured for 2 min at 90 to 95°C and separated by electrophoresis on a model ABI3730 genetic analyzer (Applied Biosystems, Foster City, CA) under conditions recommended by the manufacturer. Manual sequence editing was performed using Sequencher 4.2 (Gene Codes, Ann Arbor, MI).
Genomic analysis.
Sequences were identified by BLAST analysis (5) against sequences in the GenBank database (www.ncbi.nlm.nih.gov).
Conjugation studies.
Conjugation experiments were performed using filter mating (1). Overnight cultures of tetracycline-resistant, levofloxacin-susceptible donor (GAR3139 and GAR3141) and tetracycline-susceptible, levofloxacin-resistant recipient (GAR7071 and GAR7090) strains, grown in Luria broth, were seeded at a 1:50 dilution in separate flasks of brain heart infusion broth (BD, Sparks, MD). Following growth to early log phase (approximately 2 h) with shaking at 37°C, 5 ml each of the donor and the recipient cultures were mixed and pelleted by centrifugation (1,500 × g). The supernatant was decanted, and the pellet was carefully spread onto a nitrocellulose filter (type HA, 45-μm pore size; Millipore, Billerica, MA) and placed on a brain heart infusion broth plate. The plate was incubated for 4 hours at 37°C, after which time the filter was removed from the plate and placed in a tube with 5 ml sterile saline. The tube was vortexed in order to dislodge the cells from the filter, and serial 10-fold dilutions were prepared in sterile saline. Controls containing only the donor or the recipient were similarly processed. One hundred microliters of each dilution was spread on LB plates supplemented with 10 μg/ml tetracycline and 10 μg/ml levofloxacin. Controls were also spread onto LB agar plates containing 10 μg/ml tetracycline or 10 μg/ml levofloxacin or were not subjected to selection. Plates were incubated overnight at 37°C, and colonies were picked onto LB plates containing 10 μg/ml tetracycline and 10 μg/ml levofloxacin for further characterization. RiboPrinting was performed according to the manufacturer's instructions (Dupont-Qualicon, Wilmington, DE) to confirm the transfer of the resistance determinant from donor to recipient.
Southern blotting.
Detection of tet(M) by hybridization was carried out using a PCR digoxigenin probe and hybridization kit (Roche Molecular Systems, Summerville, NJ) according to the manufacturer's instructions. Chromosomal DNA preparations digested with AccI [single site in tet(M)] were transferred to a nylon membrane (Roche Molecular Systems). The digoxigenin-labeled ∼1.0-kb tet(M) PCR fragment (Table 2) was used as the probe. Membrane-bound DNA was hybridized at 30°C overnight, washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 68°C, developed with the alkaline phosphatase substrate CDP-Star, and visualized on X-ray film.
Cloning.
E. coli tet(M) was cloned, along with its resident promoter sequences, by using the ectet(M) clone PCR primer pair (Table 2). The amplicon was TA cloned into pCR-XL-TOPO according to manufacturer's instructions (Invitrogen, Carlsbad, CA).
Nucleotide sequence accession number.
The complete sequence of the E. coli tet(M) gene and the flanking sequences has been deposited in GenBank (accession number DQ534550).
RESULTS
Identification and cloning of E. coli tet(M).
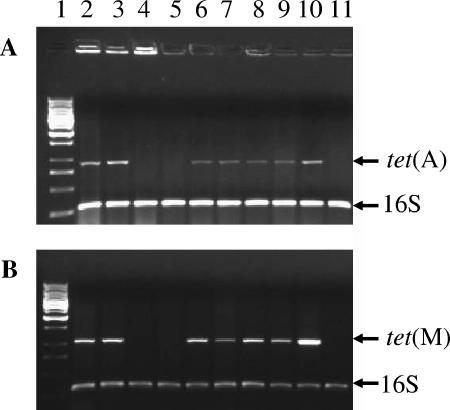
Of the 1,462 E. coli clinical isolates screened from the tigecycline phase 3 clinical trials, 333 (23%) were minocycline resistant (MIC ≥ 8 μg/ml) and 234 (16%) were tetracycline resistant (MIC ≥ 8 μg/ml) but susceptible to minocycline (MIC ≤ 4 μg/ml). Three isolates (GAR3139, GAR3141, and GAR3142), isolated from two patients from Taiwan, of the 567 screened with the tet(M) primer set resulted in products of the appropriate size (∼1 kb) (Fig. 1 and data not shown). All three strains were also positive for the tet(A) determinant.
FIG. 1.
PCR detection of tet resistance markers in E. coli clinical isolates. The PCR primer pairs for the detection of tet(A) (A) and tet(M) (B) are presented in Table 1. Templates were prepared from GAR3139 (lane 2), GAR3141 (lane 3), GAR7071 (lane 4), GAR7090 (lane 5), GC7939 (lane 6), GC7940 (lane 7), GC7941 (lane 8), GC7942 (lane 9), GC2270 as a positive control (lane 10), and DH5α (lane 11). As a control for lysate preparation, gel loading, and the PCR conditions, primers for 16S rRNA were included in the assay. Molecular weight standards were loaded in lane 1 for reference.
One strain from each patient was chosen for further study. In order to recover the 5′ and 3′ ends of the E. coli tet(M) gene, internal primers were designed from the cloned sequence and paired with corresponding upstream [ectet(M)up] and downstream [ectet(M)dwn] primers derived from the reference Staphylococcus aureus (M21136) sequence (25) (Table 2; Fig. 2). PCR resulted in products of the appropriate size, which were cloned and sequenced to complete and reconstruct the sequence of the E. coli tet(M) homologue in silico (Fig. 3). The DNA sequences from the two isolates (GAR3139 and GAR3141) analyzed differed by a single nucleotide; however, the substitution was silent (data not shown).
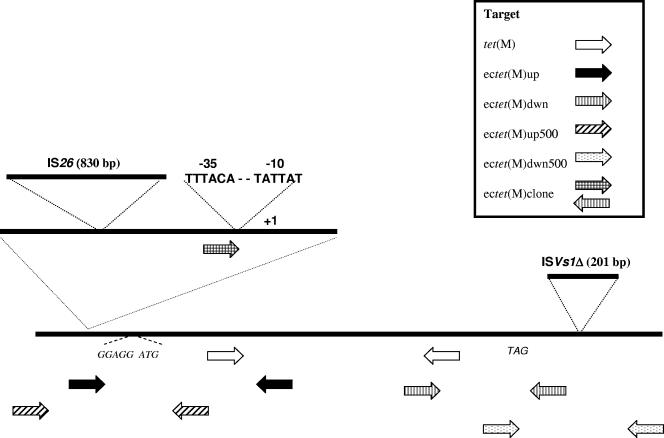
FIG. 2.
Cloning strategy and schematic diagram of the E. coli tet(M) gene and flanking regions. Locations of primers are indicated with arrows. The IS26 and ISVs1 insertions are also shown diagrammatically along with the tet(M) promoter region.
FIG.3.
Nucleotide sequence of the E. coli tet(M) gene and flanking regions. The encoded amino acid sequence of tet(M) and the putative transposase are presented by the single-letter code under the respective nucleotide sequence. The deduced amino acid sequence of the IS26 transposase tnpA encoded on the noncoding strand of IS26 is shown in italics for clarity. Transcriptional control elements for tet(M) and the start point for the Tet(M) leader peptide are indicated in boldface. The terminal repeat features for IS26 and ISVs1 are also shown in bold and underlined, and the 8-bp direct repeat sequence for IS26 is presented in bold. The silent point mutation at position 2570 that differentiates the tet(M) sequence from those of strains GAR3139 and GAR3141 is indicated in bold and underlined. DR, direct repeat; ITR-R, right inverted terminal repeat; ORF, open reading frame; ITR-L, left inverted terminal repeat; IRL, inverted repeat left.
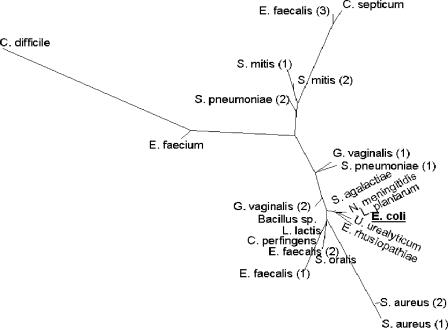
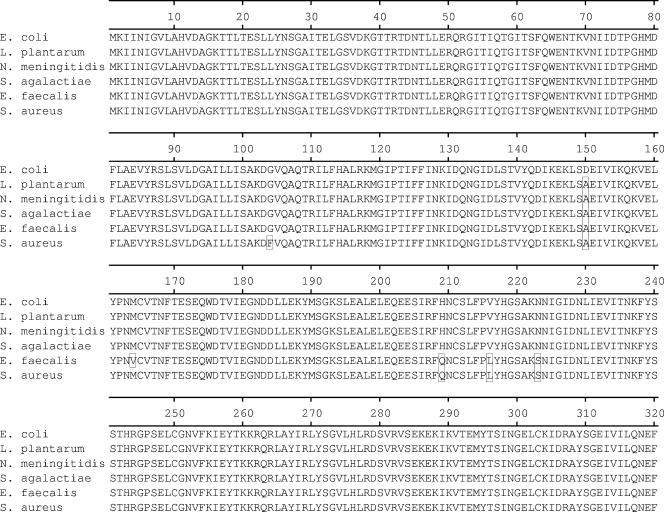
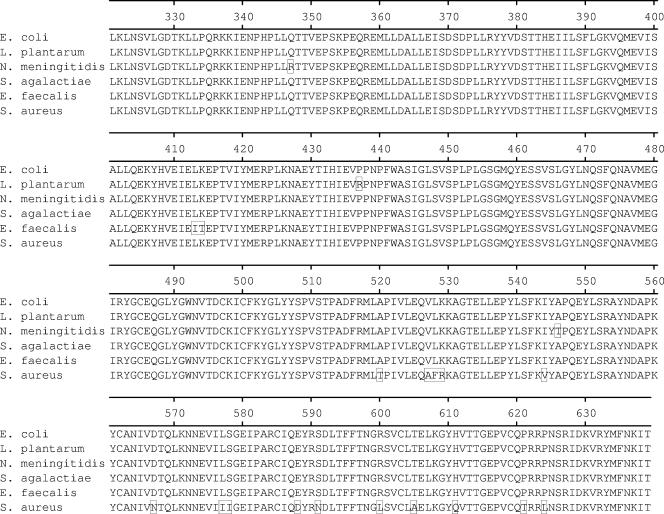
Although highly similar (approximately 90% or greater at the amino acid level) to the 23 full-length tet(M) sequences present in GenBank (www.ncbi.nlm.nih.gov), the E. coli tet(M) coding sequence was found to be most similar, ≥99.5% at the amino acid level, to the published sequences from Streptococcus agalactiae (accession no. AAM99809), Lactobacillus plantarum (AAN40886), and Neisseria meningitidis (CAA52967), differing by 1, 2, and 3 residues, respectively (15, 17, 38). The results of a phylogenetic analysis of the 23 full-length tet(M) genes and the sequence from E. coli GAR3141 are presented in Fig. 4. An amino acid alignment of the E. coli Tet(M) protein and the Tet(M) proteins from N. meningitidis (accession no. CAA52967), S. agalactiae (AAM99809), L. plantarum (AAN40886), and the S. aureus (M21136) and Enterococcus faecalis (X56353) strains are shown in Fig. 5. Compared to the S. aureus sequence used in the design of the PCR primers (25), the encoded E. coli protein differed at 20 amino acid residues.
FIG. 4.
Phylogenetic tree. An unrooted phylogenetic tree was created from a ClustalW (10) alignment of 24 unique Tet(M) protein sequences. A neighbor-joining tree was drawn by PhyloDraw 0.8 (11). The bar indicates an evolutionary distance of 0.01 amino acid substitution per position. Sequences are derived from Bacillus sp. (GenBank accession no. AAM19211), Clostridium difficile (accession no. AAO24820), Clostridium perfringens (accession no. AAK17952), Clostridium septicum (accession no. BAB71968), E. coli (this paper), E. faecalis 1 (accession no. CAA63530), E. faecalis 2 (accession no. CAA39796), E. faecalis 3 (accession no. CAA27977), Enterococcus faecium (accession no. EAN10521), Erysipelothrix rhusiopathiae (accession no. BAB82500), Gardnerella vaginalis 1 (accession no. AAB05245), Gardnerella vaginalis 2 (accession no. AAB05246), Lactococcus lactis (accession no. AAY62599), L. plantarum (accession no. AAN40886), Neisseria meningitidis (accession no. CAA52967) S. agalactiae (accession no. AAM99809), S. aureus 1 (accession no. AAA26678), S. aureus 2 (accession no. BAB56560), Streptococcus mitis 1 (accession no. CAE46077), S. mitis 2 (accession no. CAE46076), Streptococcus oralis (accession no. CAE46078), Streptococcus pneumoniae 1 (accession no. AAS45561), S. pneumoniae 2 (accession no. AAR22397), and Ureaplasma urealyticum (accession no. AAA73978).
FIG.5.
Amino acid alignment. E. coli Tet(M) was aligned with Tet(M) from L. plantarum (GenBank accession no. AAN40886), N. meningitidis (accession no. CAA52967), S. agalactiae (accession no. AAM99809), E. faecalis (accession no. CAA39796), and S. aureus (accession no. AAA26678). Amino acid residues differing from those of the E. coli protein are shown in boxes in the alignment.
Upstream sequence determination.
In order to capture upstream sequence information, we designed forward-facing primers based on Tn916 sequences located 500 and 1,000 base pairs upstream of the S. aureus coding sequence [ectet(M)up500 and ectet(M)up1000, respectively] and a reverse primer internal to the E. coli gene (Table 1; Fig. 2). Attempts to use the primer pair to clone 1,000 base pairs of upstream sequence were unsuccessful; however, the primer pair designed to clone 500 base pairs of upstream sequence resulted in a product. Interestingly, the PCR product was approximately 1.3 kb instead of the expected 500 bp, suggesting that the region upstream from the E. coli tet(M) coding sequence diverged from the published S. aureus sequence. Sequence analysis revealed that the inserted sequence was identical to the IS26 sequence encoding the transposase gene tnpA, initially described by Mollet et al. (22). The IS26 element inserted 113 bp upstream of the −35 element of the tet(M) promoter (25, 37). The promoter elements (−10, −35, +1 start site) of the tet(M) gene as well as the translational start site and putative regulatory control region within the leader peptide are completely conserved in E. coli, by comparison to the sequence published by Nesin et al. (25) (Fig. 3).
Downstream sequence determination.
Downstream sequence information was obtained using reverse-facing primers based on Tn916 sequences 500 and 1,000 base pairs downstream of the S. aureus tet(M) coding sequence [ectet(M)dwn500 and ectet(M)dwn1000, respectively] and a forward-facing primer internal to the E. coli gene (Table 1; Fig. 2). Again, the attempt to clone the larger downstream fragment using the Tn916 primer 1 kb from the end of the coding sequence was unsuccessful. However, the use of the ectet(M)dwn500 primer resulted in a fragment approximately 700 bp in length that, upon sequencing, revealed a 200-bp insertion sequence that was identical to a sequence previously reported to occur in the V. salmonicida plasmid pRVS1 and referred to as ISVs1 (36). The ISVs1 sequence inserted 170 bp downstream of the tet(M) stop codon (Fig. 3). The downstream sequence encoding the putative tet(M) transcriptional terminator was not captured for analysis.
Conjugation studies.
It was of some interest to determine if the E. coli tet(M) gene was associated with a mobile element, as the sequence analysis revealed that at least two insertion sequences were located proximal to the gene. As shown in Table 3, resistance to both tetracycline and minocycline was successfully transferred by conjugation from both GAR3139 and GAR3141 to two tetracycline-susceptible recipient strains (GAR7071 and GAR7090). Ribotyping was used to confirm that the resistance determinants moved from the donor to the recipient (data not shown). In both cases, PCR analysis revealed that both the E. coli tet(M) and the tet(A) gene were mobilized into the donor. Analysis by direct PCR and Southern blotting using a PCR amplicon [tet(M) forward and reverse primers (Table 2)] as the probe demonstrated the presence of the E. coli tet(M) gene in the recipient strains (data not shown). MIC analysis of the recipient strains (Table 3) indicated that the tet(M) gene is functional in the transconjugants, as tet(A) does not efficiently efflux minocycline (28).
TABLE 3.
Susceptibility data
| E. coli strain | Tet resistance determinant(s)a | MIC (μg/ml)b
|
|||
|---|---|---|---|---|---|
| Tetracycline | Minocycline | Levofloxacin | Tobramycin | ||
| GAR3139 | tet(A), tet(M) | >64 | 8 | 0.25 | 0.5 |
| GAR3141 | tet(A), tet(M) | >64 | 4 | 0.5 | 0.5 |
| GAR7071 | None | 2 | 1 | >16 | 8 |
| GAR7090 | None | 4 | 4 | >16 | 1 |
| GC7941 | tet(A), tet(M) | >64 | 16 | >16 | 4 |
| GC7942 | tet(A), tet(M) | >64 | 16 | >16 | 0.5 |
| GC7949 | tet(M) | >64 | 32 | ND | ND |
| PCR-XL-TOPO | None | 1 | 0.5 | ND | ND |
Genes were detected by PCR except for the GC7949 strain, which contains tet(M) cloned into pCR-XL-TOPO.
ND, not done.
Cloning and expression of E. coli tet(M).
The E. coli tet(M) gene was cloned, along with its resident promoter, from the clinical isolate and expressed in E. coli. As shown in Table 3, the cloned E. coli-derived tet(M) gene conferred resistance to tetracycline and minocycline on the recombinant E. coli host strain.
DISCUSSION
The first tetracycline, chlortetracycline (Aureomycin), was identified and developed through the ingenuity and dedication of Benjamin Duggar and Lederle Laboratories in the late1940s (16). Tetracycline resistance mechanisms appeared soon after this antibiotic class was introduced into the marketplace in 1948. Although the efflux mechanism of tetracycline resistance quickly appeared in enteric pathogens (2), the ribosomal protection mechanism of resistance had not been detected in enteric organisms until 2004 (8). This is curious as both resistance mechanisms are associated with mobile elements and the tet(M) gene has been detected in other gram-negative hosts, such as Neisseria, Haemophilus spp., and Campylobacter spp.(12).
We report the identification tet(M) in clinical isolates of E. coli from two patients enrolled in phase 3 clinical trials for the recently approved broad-spectrum antibiotic tigecycline (6, 26). Both patients were undergoing treatment for complicated intra-abdominal infections at the same hospital in Taiwan. The tet(M) genes from the clinical isolates were most similar to tet(M) genes from S. agalactiae, L. plantarum, and N. meningitidis.
Based on the premise that the source of the E. coli tet(M) gene was the Tn916/Tn1545 family of conjugative transposons (13), PCR primers were designed based on Tn916 sequences located upstream and downstream of the tet(M) open reading frame in order to recover the entire gene and flanking regions. The fact that this approach worked indicated that Tn916 sequences were contained in the element that mobilized into E. coli. However, the identification of additional insertion sequences identical to IS26 and to ISVs1 suggested that there may have been one or more intermediate hosts between the original host and the clinical E. coli strains. Preliminary experiments using PCR primers 1,000 bp upstream and downstream of the tet(M) structural gene to recover additional sequences were unsuccessful, suggesting that only limited Tn916-derived sequences remain associated with the tet(M) gene in E. coli.
The IS26 insertion sequence was first described in association with Tn2680, which encodes kanamycin resistance on the R plasmid, Rts1, from Proteus vulgaris (22). IS26 is 820 bp long and carries 14-bp perfect inverted terminal repeats flanking the tnpA gene (20). Upon integration, IS26 creates an 8-bp duplication at the insertion site; the sequence ATTTGATT is found as a direct repeat flanking the IS26 inverted terminal repeat at the insertion site of the element upstream of the tet(M) promoter in E. coli. The ISVs1 insertion sequence has been described only in association with a 170-MDa resistance plasmid, pRVS1, which encodes a Tet(E) homologue in V. salmonicida (36). The sequence of ISVs1 deposited in GenBank encodes a 96-residue truncated transposase gene product, whereas the sequence associated with tet(M) encodes the C-terminal 36 residues of the transposase. The sequence in the present report also encodes the complete left inverted terminal repeat region of 43 base pairs.
The IS26 element inserted 113 bp upstream of the tet(M) promoter sequence. All upstream transcriptional and translational control sequences were maintained intact in the E. coli gene compared to that reported by Nesin et al. (25). Due to the inability to amplify downstream sequences beyond the ISVs1 insertion, we were unable to confirm the presence of the tet(M) terminator region reported by previous investigators (37).
We propose that the E. coli tet(M) gene originated in a Tn916 host, most likely a streptococcal species, although the limited flanking sequence data indicate that most Tn916 sequences have been lost. As ISVs1 insertion element sequences have not, until this report, been reported to occur in any organism except V. salmonicida (36), we further propose that the element passed through V. salmonicida and additional gram-negative hosts, acquiring IS26 before moving into E. coli.
In order to get an unbiased view of the presence and types of tet genes in natural (nonclinical) nonselected populations of bacteria in the environment, Bryan et al. (8) screened 1,263 isolates from 12 animal sources and humans for 14 tetracycline resistance genes. The investigators report the presence of tet(M) in a number of isolates sourced from both pigs and chickens. A partial sequence, 386 bp, of the tet(M) amplicon was found to be 98% identical to the tet(M) gene from E. faecalis (8). As the complete sequence, including flanking regions, was not reported nor submitted to GenBank, a comparison to the sequences from human clinical isolates was not possible.
The work of a number of investigators (8, 19) suggests that the environmental exposure of humans and animals to tetracyclines and other antibiotics drives the development and dissemination of resistance determinants by horizontal gene transfer. Nevertheless, until recently, the tet(M) tetracycline resistance determinant had not been reported for E. coli. This is despite evidence that cloned tet(M) genes from S. aureus and E. faecalis have been found to be expressed and functional in E. coli and the demonstration of the in vivo transfer of Tn916 from E. faecalis to E. coli (29). Our data support the recent report (8) of tet(M) in E. coli isolates from farm animals and extend the finding that the ribosomal protection mechanism of tetracycline resistance, mediated by tet(M), has migrated into human clinical isolates of E coli.
These findings uphold the continued transfer of antibiotic resistance determinants among various environmental and clinical bacterial populations, which is a motivating factor for the ongoing search for novel antibacterial agents in the age of resistance. As a response to the diminished utility of the tetracyclines (12, 32, 33), the novel glycylcycline agent tigecycline was recently brought to the marketplace and has shown potent in vitro activity against tetracycline- and minocycline-resistant strains (7, 28).
Acknowledgments
We thank Steve Projan, Alexey Ruzin, and Peter Petersen for critical reading of the manuscript. We also thank Guy Singh for RiboPrinting strains and Jan Kieleczawa and the Wyeth Core Sequencing facility for sequence analysis.
REFERENCES
- 1.Agersø, Y., and D. Sandvang. 2005. Class 1 integrons and tetracycline resistance genes in Alcaligenes, Arthrobacter, and Pseudomonas spp. isolated from pigsties and manured soil. Appl. Environ. Microbiol. 71:7941-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiba, T., K. Koyama, Y. Ishiki, S. Kimura, and T. Fukushima. 1960. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn. J. Microbiol. 4:219-227. [DOI] [PubMed] [Google Scholar]
- 3.Allard, J. D., and K. P. Bertrand. 1993. Sequence of a class E tetracycline resistance gene from Escherichia coli and comparison of related tetracycline efflux proteins. J. Bacteriol. 175:4554-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allard, J. D., M. L. Gibson, L. H. Vu, T. T. Nguyen, and K. P. Bertrand. 1993. Nucleotide sequence of class D tetracycline resistance genes from Salmonella ordonez. Mol. Gen. Genet. 237:301-305. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babinchak, T., E. J. Ellis-Grosse, N. Dartois, G. M. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline in the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41:S354-S367. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, P. A., D. T. Weaver-Sands, and P. J. Petersen. 2005. In vitro activity of tigecycline against isolates from patients enrolled in phase 3 clinical trials for complicated skin and skin structure infections and complicated intra-abdominal infections. Clin. Infect. Dis. 41(Suppl. 5):S315-S332. [DOI] [PubMed] [Google Scholar]
- 8.Bryan, A., N. Shapir, and M. J. Sadowsky. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burdett, V. 1986. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J. Bacteriol. 165:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J. H., H. Y. Jung, H. S. Kim, and H. G. Cho. 2000. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics 16:1056-1058. [DOI] [PubMed] [Google Scholar]
- 12.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement, vol. 25. CLSI/NCCLS M100-S15. CLSI, Wayne, Pa.
- 15.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 16.Duggar, B. M. 1948. Aureomycin: a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 51:171-181. [DOI] [PubMed] [Google Scholar]
- 17.Gascoyne-Binzi, D. M., J. Heritage, P. M. Hawkey, and M. S. Sprott. 1994. Characterization of a tet(M)-carrying plasmid from Neisseria meningitidis. J. Antimicrob. Chemother. 34:1015-1023. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S. B. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33(Suppl. 3):S124-S129. [DOI] [PubMed] [Google Scholar]
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, P., P. Trieu-Cuot, and P. Courvalin. 1986. Nucleotide sequence of the tet(M) tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 14:7047-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollet, B., S. Iida, J. Shepherd, and W. Arber. 1983. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 11:6319-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCCLS. 2003. Methods for dilution antimicrobial disk susceptibility tests, 8th ed., vol. 20. Approved standard M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed., vol. 23. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Nesin, M., P. Svec, J. R. Lupski, G. N. Godson, B. Kreiswirth, J. Kornblum, and S. J. Projan. 1990. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2273-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliva, M. E., A. Rekha, A. Yellin, J. Pasternak, M. Campos, G. M. Rose, T. Babinchak, E. J. Ellis-Grosse, and E. Loh for the 301 Study Group. 2005. A multicenter trial of the efficacy and safety of tigecycline versus imipenem/cilastatin in patients with complicated intra-abdominal infections. BMC Infect. Dis. 5:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peden, K. W. 1983. Revised sequence of the tetracycline-resistance gene of pBR322. Gene 22:277-280. [DOI] [PubMed] [Google Scholar]
- 28.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antimicrobial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poyart, C., J. Celli, and P. Trieu-Cuot. 1995. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob. Agents Chemother. 39:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, M. C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36:462-467. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, M. C., Y. Pang, D. E. Riley, S. L. Hillier, R. C. Berger, and J. N. Krieger. 1993. Detection of tet(M) and tet(O) tetracycline resistance genes by polymerase chain reaction. Mol. Cell. Probes 7:387-393. [DOI] [PubMed] [Google Scholar]
- 35.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sørum, H., M. C. Roberts, and J. H. Crosa. 1992. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob. Agents Chemother. 36:611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su, Y. A., P. He, and D. B. Clewell. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyeth Pharmaceuticals. 2005. Tygacil, package insert. Wyeth Pharmaceuticals, Inc., Collegeville, Pa.
- 40.Yang, W., I. F. Moore, K. P. Koteva, D. C. Bareich, D. W. Hughes, and G. D. Wright. 2004. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279:52346-52352. [DOI] [PubMed] [Google Scholar]