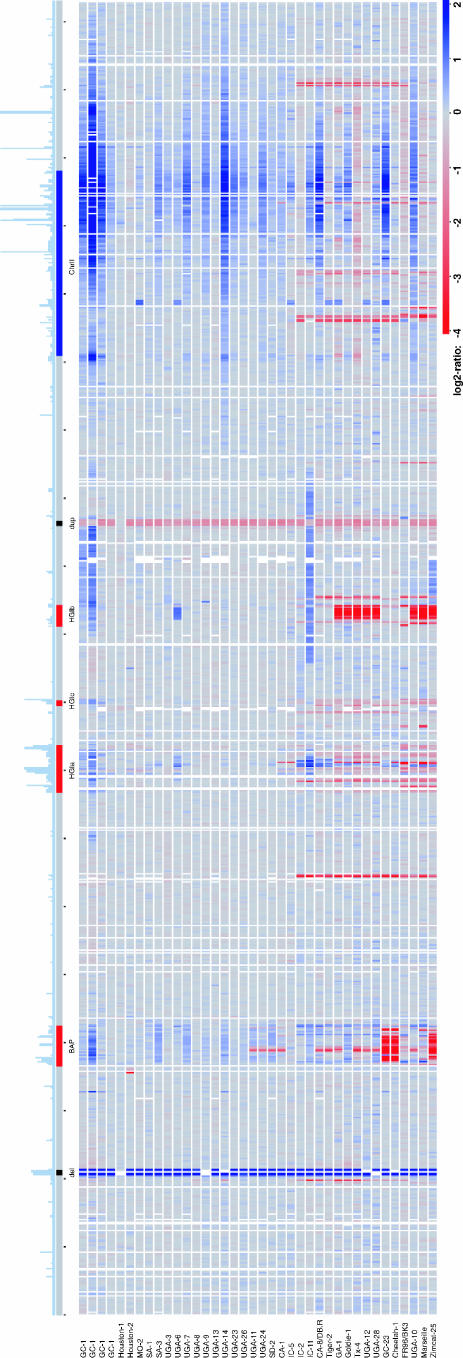
FIG. 1.
Linear representation of the microarray CGH data ordered according to position in the Houston-1Seq genome. The frequency of repeats in the Houston-1Seq genome is shown to scale above the genome at the top (the number at which each sequence occurs in the genome with more than 80% sequence identity over 100 bp). The positions of the prophage region (BAP) and the genomic islands (HGIa, HGIb, and HGIc) in the Houston-1Seq genome are shown in red, and the chromosome II-like region (ChrII) is shown in blue. The regions deleted (del) and duplicated (dup) in the Houston-1Ref strain, used as a reference in the hybridization experiments, are indicated in black. Each horizontal line below the Houston-1Seq genome represents an isolate, with the color corresponding to the hybridization signal for the isolate relative to the Houston-1Ref strain (red for lower and blue for higher signal).