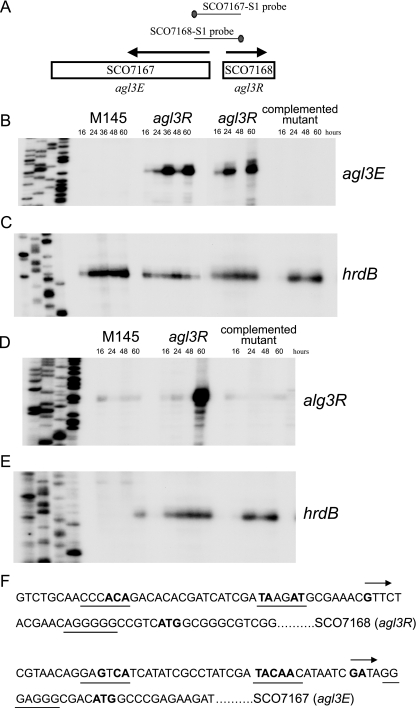
FIG. 3.
S1 nuclease mapping of transcripts originating upstream of agl3R and agl3E. RNA was isolated from the wild type (M145), the agl3R mutant, and the agl3R mutant containing a wild-type copy of the agl3R gene. Panel A shows the position of the probe used for the detection of agl3R and agl3E transcripts. Panel B shows the protected RNA of a transcript originating upstream of agl3R isolated from the wild type, the agl3R mutant (two different experiments), and the agl3R mutant containing the agl3R wild-type allele. Panel D shows the protected RNA of a transcript originating upstream of agl3E isolated from the wild type, the agl3R mutant, and the agl3R mutant containing the agl3R wild-type allele. Panels C and E show transcripts originating upstream of the hrdB gene as a control for RNA. In all cases, the sequencing reactions were loaded A, G, C, and T. (F) DNA sequence upstream of the apparent transcription start sites (indicated by boldface type and an arrow) for agl3R and agl3E. The potential −35 and −10 region RNA polymerase recognition sequences and bases with consensus to TTGACA and TACAAT, prototypical of vegetative promoters of Streptomyces, are in boldface type and underlined. Putative ribosome binding sites are underlined, and the annotated translation start sites (ATG for each) are also in boldface type.