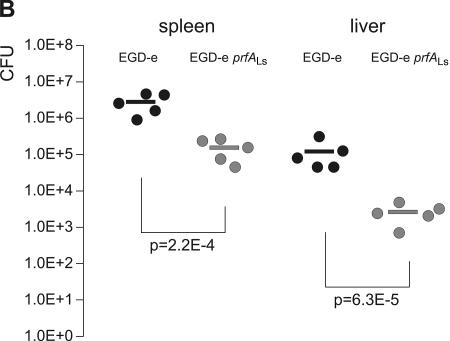
FIG. 10.
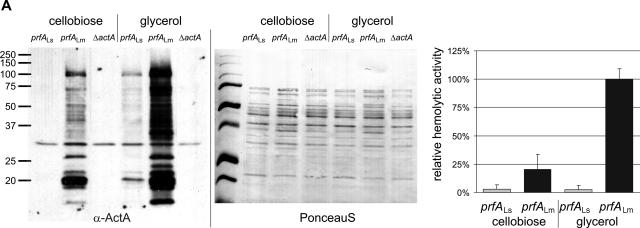
Allelic replacement of prfALm with prfALs in Listeria monocytogenes. (A) Expression of the PrfA-regulated genes actA and hly in the strains L. monocytogenes EGD-e prfALs and L. monocytogenes EGD-e prfALm. The strains were grown to an optical density at 600 nm of 0.6 in minimal medium supplemented with 50 mM cellobiose or glycerol. Cytoplasmic proteins were prepared, and equal amounts (5 μg) were separated by SDS-PAGE and analyzed by Western blotting. After use of a loading control with PonceauS, immunodetection of ActA protein (α-ActA) was performed. Protein preparations of the strain L. monocytogenes EGD-e ΔactA (S. Pilgrim and S. Bauer, unpublished data) are also shown as a negative control. The hemolytic activities of L. monocytogenes EGD-e prfALs and L. monocytogenes EGD-e prfALm grown in minimal medium to an optical density at 600 nm of 0.6 are shown to the right. The hemolytic activity was determined in three independently performed experiments; the error bars indicate standard deviations of the means for the three experiments. (B) Viable bacterial counts in spleens and livers of C57BL/6 mice infected intravenously with 5 × 103 CFU of wild-type L. monocytogenes EGD-e or L. monocytogenes EGD-e prfALs (EGD-e s-prfA). Bacterial loads in spleens and livers of infected animals are shown. Each symbol represents a single animal. The lines indicate the means for the experimental groups (n = 5). For each organ, the P value determined with Student's t test of the log-transformed CFU amounts is given. L. monocytogenes EGD-e prfALs was highly significantly (P ≤ 0.01) attenuated compared to the wild-type strain.