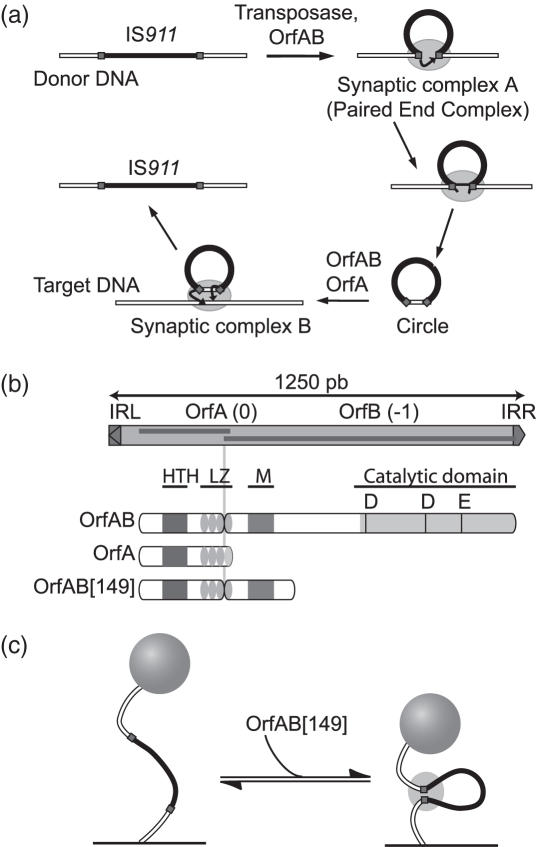
Figure 1.
The IS911 transposition cycle, organization of its proteins and single-molecule application. (a) Steps in IS911 transposition. IRs: squares; IS911 sequence: black box; transposase OrfAB: ellipse. The strand cleavage and transfer reactions are indicated by arrows. The transposase stoichiometry in these synaptic complexes is not known. The PEC and circle transposition intermediates are described in the text. (b) Organization of IS911 and its protein products (OrfA and the transposase OrfAB). IRs: triangles; open reading frames (orf): dark boxes. The DNA binding domain is composed of a Helix–Turn–Helix motif (HTH, grey box), a Leucine Zipper (LZ, ellipses) and a multimerization domain (M, grey box). The catalytic domain carries a classical DDE motif. The protein used in the assay, OrfAB[149], is a derivative of the OrfAB transposase amputated for the C-terminal catalytic DDE domain. (c) Principle of the TPM method. Labelled DNA (white line) with IS911 sequence (black line) and two IRs (squares) is tethered at one end to a glass surface and a polystyrene bead is attached to the other. Looping of DNA by OrfAB[149] (ellipse) decreases the effective tether length and the amplitude of the bead motion.