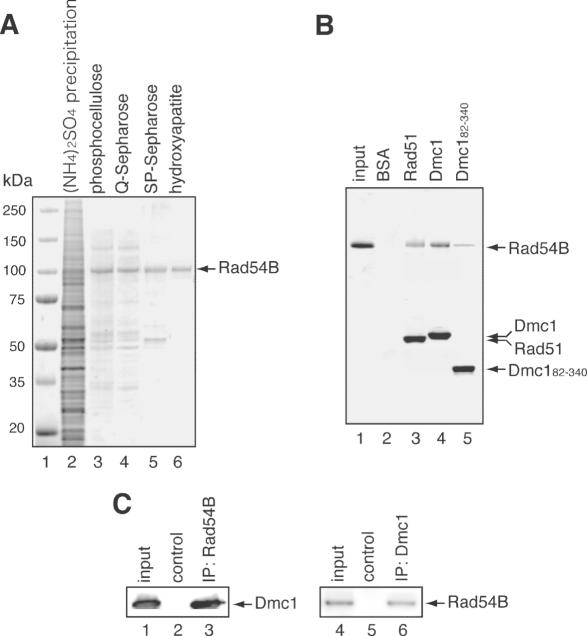
Figure 1.
Rad54B purification and complex formation. (A) SDS–PAGE of the column fractions containing Rad54B. The proteins precipitated by ammonium sulfate (lane 2), the phosphocellulose peak fraction (lane 3), the Q-Sepharose flow-through (lane 4), the SP–Sepharose peak fraction (lane 5) and the hydroxyapatite peak fraction (lane 6) were fractionated on a 4–20% SDS–PAGE gel and stained with Coomassie brilliant blue. Lane 1 indicates the molecular mass markers. (B) Protein–protein interaction assay of Rad54B with Rad51, Dmc1 and Dmc182–340. Rad54B was mixed with either Rad51, Dmc1 or Dmc182–340 that was covalently conjugated to an Affi-Gel 15 matrix. After an incubation at room temperature, the Affi-Gel 15 matrices were washed with binding buffer, directly mixed with 2-fold SDS–PAGE sample buffer and fractionated on a 4–20% gradient SDS–PAGE gel. Lane 1 is one-tenth (2 µg) of the input proteins, lane 2 is the negative control using the Affi-Gel 15 matrix conjugated with BSA, lane 3 is the Affi-Gel 15 matrix conjugated with Rad51, lane 4 is the Affi-Gel 15 matrix conjugated with Dmc1 and lane 5 is the Affi-Gel 15 matrix conjugated with Dmc182–340. The bands were visualized by Coomassie brilliant blue staining. Notably, the Rad51, Dmc1 and Dmc182–340 proteins form multimers and thus, these proteins can indirectly associate with the Affi-Gel matrix via interactions with proteins crosslinked to the matrix. Rad51, Dmc1 and Dmc182–340 detected in lanes 3–5 represent fractions that were not covalently conjugated to the Affi-Gel beads. In contrast, BSA was not detected (lane 2), since BSA does not form multimers, and all proteins were covalently conjugated to the beads. (C) Immunoprecipitation (IP) experiments of Rad54B and Dmc1. Sf9 insect cells were infected with a baculovirus containing both the Dmc1 and Rad54B genes. The cells were lysed and subjected to immunoprecipitation with anti-Rad54B antibody (lane 3) or anti-Dmc1 antibody (lane 6). The precipitates were analyzed by immunoblotting. As negative control experiments, the cell lysate was incubated with normal IgG (lanes 2 and 5). Lanes 1 and 4 are one-tenth of the input cell lysate.