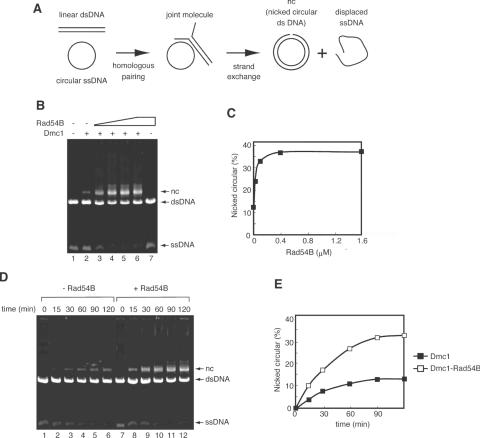
Figure 2.
Stimulation of the Dmc1-mediated DNA strand exchange activity by Rad54B. (A) Schematic representation of the DNA strand exchange assay. The nucleoprotein filament is formed by incubating Dmc1 with circular ssDNA. The filament is paired with homologous linear dsDNA to yield a joint molecule. A nicked circular duplex (nc) and linear ssDNA are then generated by completing strand exchange over the length of the DNA molecules. (B) Dmc1-mediated DNA strand exchange activity with Rad54B. A constant amount of Dmc1 (7.5 µM) was incubated for 2 h with φX174 circular ssDNA (30 µM), φX174 linear dsDNA (22 µM), RPA (2 µM), KCl (200 mM) and increasing concentrations of Rad54B (0, 0.025, 0.1, 0.4 and 1.6 µM in lanes 2–6, respectively), in the order described for the procedure at 37°C. In lane 1, ssDNA and dsDNA were incubated in buffer with RPA and KCl, but without other recombinant proteins, and in lane 7, ssDNA and dsDNA were incubated in buffer with RPA, KCl and Rad54B (1.6 µM), but no Dmc1. The reaction mixtures were deproteinized, fractionated on a 1% agarose gel and stained with ethidium bromide. (C) Graphic representation of the experiments shown in (B). The amounts of nicked circular duplex are presented. (D) Time course experiments for DNA strand exchange. Dmc1 (7.5 µM) was incubated with φX174 circular ssDNA (30 µM), φX174 linear dsDNA (22 µM), RPA (2 µM) and KCl (200 mM), in the presence (open squares) or absence (closed squares) of Rad54B (0.4 µM), in the order described for the procedure at 37°C, for the indicated times. (E) Graphic representation of the experiments shown in (D). The amounts of nicked circular duplex are graphically presented.