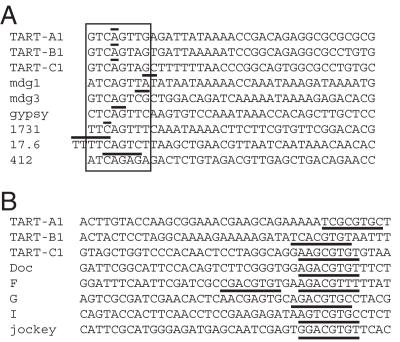
Figure 4.
Alignment of TART transcription initiation sites with initiation sites of other Drosophila retrotransposons. (A) The 5a end of each TART subfamily is aligned with the 5′ start sites of several D.melanogaster LTR retrotransposons. An 8 nt sequence that is a close match to a consensus D.melanogaster initiator sequence, A/G/T-T-C-A-G/T-T-C/T-G (27), is boxed in all of the sequences. Experimentally determined transcription initiation sites are indicated with horizontal lines above one or more bases; for the TART sequences only the most common start site is indicated (bold positions in Table 2). Each sequence begins with the initiator consensus or the transcription initiation site. References for these sites are: mdg1, mdg3 and gyspy (40), 1731 (41), 17.6 (42) and 412 (43). (B) The 5d end of each TART subfamily is aligned with the 5′ start sites of several D.melanogaster non-LTR retrotransposons. Each alignment begins with the transcription initiation site. The corresponding sites for TART-A1, -B1 and -C1 are 12 645, 9749 and 10 133, respectively and the sequences shown are the antisense strand. As described in the text, the start site for TART-C1 end 5d was taken from an EST sequence in the GenBank database. Underlined sequences match an extended version of a downstream promoter element (13,17,44). References for the other non-LTRs are: Doc (13), F (10), G (17,45), I (11) and jockey (8).