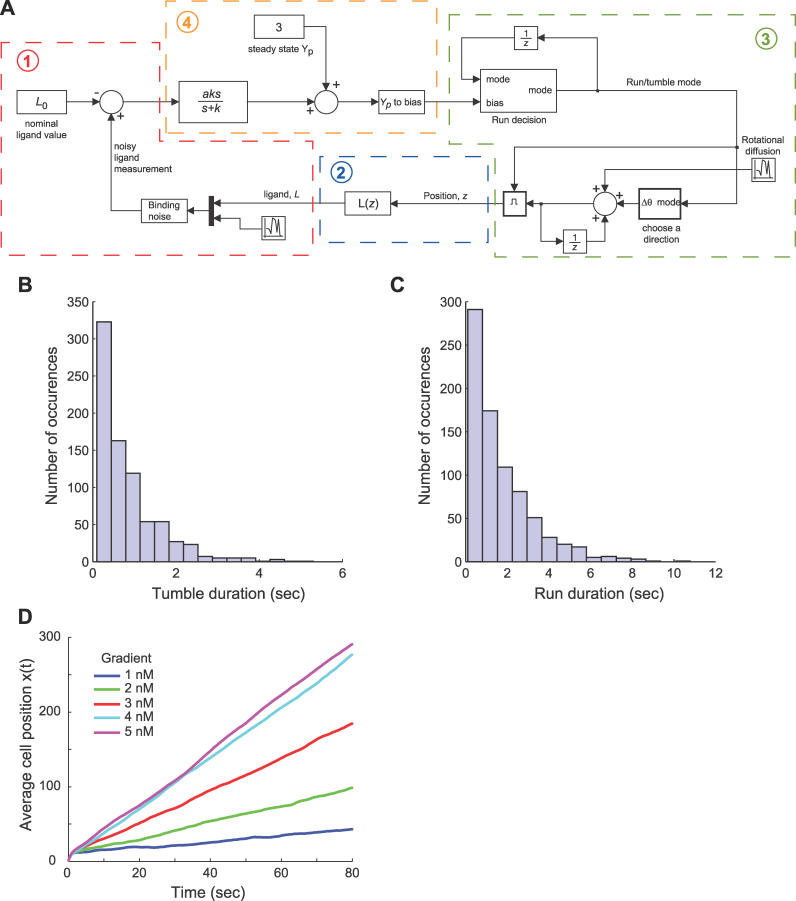
Figure 11. Model of Bacterial Chemotaxis.
(A) The chemotaxis simulation assumes the following feedback interaction between the bacteria and its environment: 1) the external environment affects the sensing model through the external ligand concentration L and incorporates the effect of binding noise; 2) ligand concentration is determined by the location of the cell in the environment (the spatial location z); 3) spatial location is determined by integrating the velocity and angle, which incorporates the effect of rotational diffusion and the “run/tumble” decisions; and 4) the run/tumble decisions are based on the signaling mechanism's response (that is, the filter + differentiation) to the measured ligand concentration.
(B,C) Validation of chemotaxis model histograms of tumble (B) and run (C) durations for an unstimulated (g = L 0 = 0) E. coli with a simulation time of 2,000 s. Both distributions exhibit characteristics of a Poisson process.
(D) Average simulation results of 500 runs of a bacteria swimming in a gradient of ligand concentration with varying chemoattractant slopes.