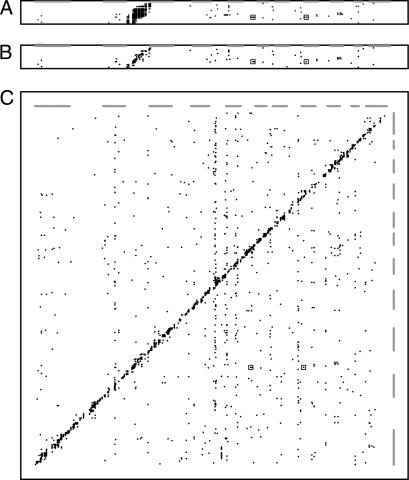
Fig. 1.
Maps of eQTLs observed in the rat eye. Expressed transcripts are represented in genome order on the y axis, whereas genetic markers are represented in genome order on the x axis. (A) Linkage data from transcripts on chromosome 4. The relative statistical significance of the relationship between a given marker and a given transcript is shown with different densities of the plotted points (black = α of 0.00001; lightest gray = α of 0.05). The boxes indicate the locations of data corresponding to the significant noncontiguous linkages to the Opn1sw transcript. (B) Linkage data from chromosome 4 but with redundant linkages (multiple genetic markers detecting the same eQTL) removed as described. In addition, all linkages that are significant at an α level of 0.05 are shown with symbols of identical size and density. (C) Linkage data from the entire genome shown in the same fashion as in B. The genomic positions of the odd-numbered chromosomes are depicted as gray bars along both axes.