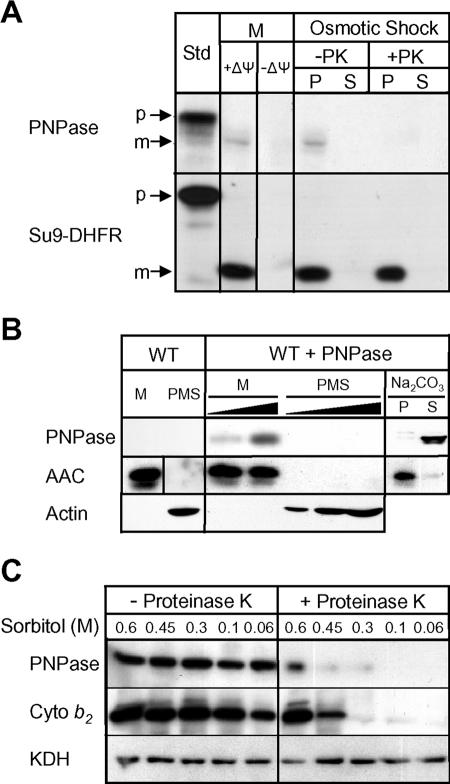
FIG. 3.
PNPase is actively imported and processed and localizes to the mitochondrial IMS. (A) Radiolabeled PNPase was imported into isolated yeast mitochondria (M) at 25°C in the presence and absence of Δψ. The OM was ruptured by osmotic shock to generate mitoplasts in the presence and absence of proteinase K (PK). Centrifugation separated mitoplasts (P) from the soluble IMS fraction (S). As a control, matrix-localized Su9-DHFR was also imported. Samples were analyzed by SDS-PAGE and fluorography. Standard (Std) refers to 10% of the radioactive precursor added to each assay. p, precursor; m, mature. (B) PNPase was expressed in S. cerevisiae under the control of the regulated CUP1 promoter (WT + PNPase). Cells were fractionated and mitochondria (M) were separated from the postmitochondrial supernatant (PMS) by centrifugation. Mitochondria were incubated with 0.1 M Na2CO3 (pH 11.5), and integral membrane proteins (P) were separated from soluble proteins (S) by centrifugation. Fifty- and 100-μg portions of M and increasing amounts of PMS were loaded in adjacent lanes. PNPase, the IM marker AAC protein, and the cytosolic marker β-actin were detected by immunoblotting. WT, wild type. (C) Yeast mitochondria containing PNPase as described for panel B were subjected to osmotic shock (incubation in solution with decreasing sorbitol concentration) in the presence and absence of proteinase K. Mitoplasts were recovered by centrifugation and separated by SDS-PAGE. PNPase, IMS marker cytochrome b2, and matrix marker KDH were detected by immunoblotting.