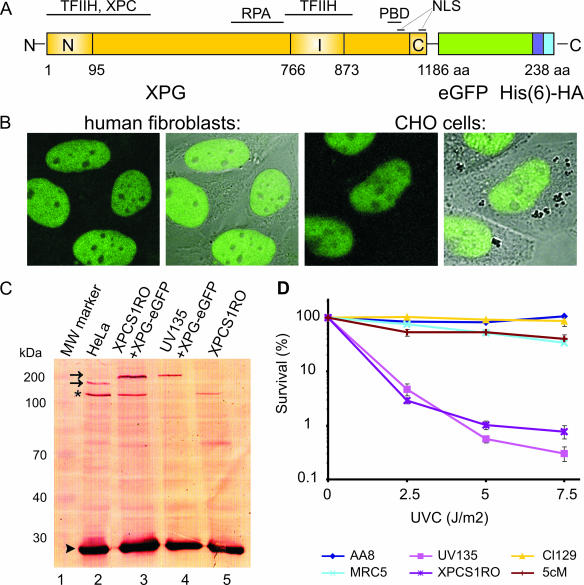
FIG. 1.
Expression and functionality of XPG-eGFP. (A) Schematic representation of the XPG-eGFP-His6-hemagglutinin (HA) fusion gene, with the N-terminal and C-terminal nuclease domains (N and C, respectively) and different interaction domains indicated. I, internal domain; PBD, PCNA binding domain; NLS, probable nuclear localization signal; aa, amino acids. (B) Localization of the XPG fusion protein in human fibroblasts (two images at left, showing the fluorescence signal and an overlay of fluorescence and phase contrast) and CHO cells (two images at right). XPG-eGFP is present mainly in the nucleus, except in the nucleoli. (C) Immunoblot (probed with monoclonal anti-XPG) of 40 μg of whole-cell extract from HeLa cells (lane 2), human XPCS1RO-Sv (XP-G) cells expressing XPG-eGFP (lane 3), CHO (UV135) cells expressing XPG-eGFP (lane 4), and untransfected XPCS1RO-Sv cells (lane 5). The molecular masses of protein markers are indicated in kilodaltons (kDa). eGFP-tagged XPG migrates slower than endogenous XPG (upper and lower arrows, respectively). No XPG protein was detected in the human fibroblasts, in which the severely truncated XPG-mRNA was probably highly unstable or not recognized. Chinese hamster ovary cell XPG was not detected with our anti-XPG serum. Loading control: PCNA (arrowhead). The asterisk indicates a cross-reacting nonspecific band only present in human cell extracts. (D) UV survival of repair-proficient human MRC5 cells (wild type; light blue line), XPCS1RO cells (violet line), XPCS1RO cells stably expressing XPG-eGFP (clone 5 cM; brown line), wild-type CHO cells (AA8; dark blue line), XPG-deficient CHO cells (UV135; purple line), and UV135 cells expressing XPG-eGFP (clone 129; yellow line). The transfected cell lines show a correction of UV sensitivity to the wild-type level.