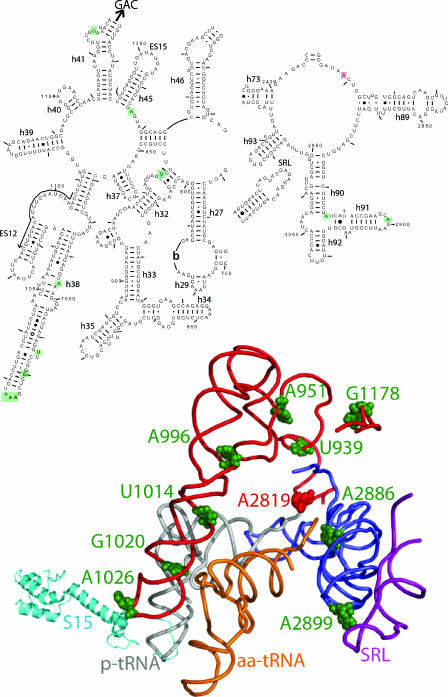
FIG. 7.
An arc of unpaired “hinge bases” facilitate information exchange among functional centers of the ribosome. The illustration at the top of the figure is a two-dimensional map showing the regions of yeast 25S rRNA of interest. Bases boxed in green show enhanced in-line cleavage in the 25S rRNA mutants. A2819 in the core of the peptidyltransferase center is blocked in red. Helices and expansion segments are indicated. The locations of every 50th base are labeled, and every 10th base is indicated by tick marks. The arrow indicates the direction to the GTPase-associated center (GAC). The lower diagram shows the positions of bases of interest modeled onto the atomic resolution structure of the E. coli ribosome (39). The labeling follows the conventions described in Fig. 2. The peptidyl-tRNA (p-tRNA) is gray, aa-tRNA is orange, the helices 32, 37, 38, and 41 are red, helix 91 is blue, and the sarcin/ricin loop (SRL) is purple. Hinge bases are shown as green spheres, and A2819 is shown in red.