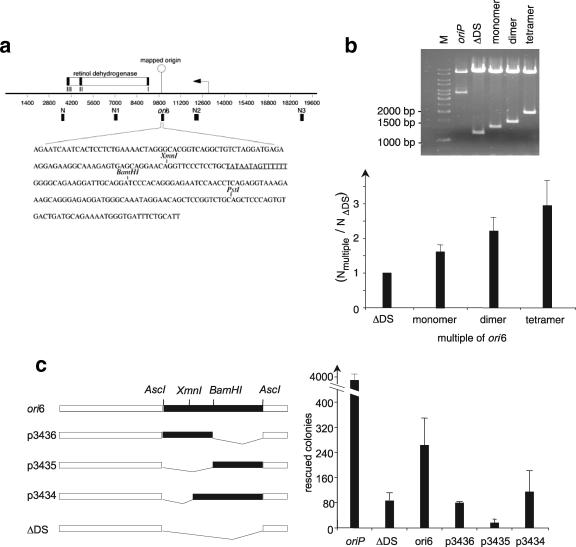
FIG. 4.
Genetic analysis of ori6. (a) The Orc2-precipitated ori6 sequence is located in the 5′ untranslated region of the retinol dehydrogenase gene. The genomic organization of this region is shown on top. The arrowhead depicts the transcription start site, and the putative exons are shown as black boxes (I to III). The location of the cloned Orc2-binding fragment is shown below the line (ori6), as are the locations of the control regions amplified by quantitative PCR (N to N3). The 249-bp-long fragment, including the restriction enzymes used for the deletion mutants as well as an AT-rich region (underlined), is depicted below. (b) ori6 fragments were multimerized in a head-to-tail fashion. The ori6 sequence was PCR amplified from the ori6 plasmid with one primer containing an AscI restriction site and one containing an AflIII restriction site. After the PCR, ori6 fragments were ligated to each other by a head-to-tail cloning strategy and ligated into plasmid p2932 (ΔDS). Replication competence was determined as described above in three independent transient replication assays. (c) Deletion analysis of ori6 located at the retinol dehydrogenase gene. Different deletion mutants of ori6 are depicted on the left. Using the indicated internal restriction sites, XmnI and BamHI, generated the deletion mutants. The replication competences of the different mutants were determined in transient replication assays with HEK293/EBNA1 cells and quantified by counting the rescued colonies. The histogram shows the average values and standard deviations for triplicates.