Abstract
Pandemic influenza remains a serious public health threat and the processes involved in the evolutionary emergence of pandemic influenza strains remain incompletely understood. Here, we develop a stochastic model for the evolutionary emergence of pandemic influenza, and use it to address three main questions. (i) What is the minimum annual number of avian influenza virus infections required in humans to explain the historical rate of pandemic emergence? (ii) Are such avian influenza infections in humans more likely to give rise to pandemic strains if they are driven by repeated cross-species introductions, or by low-level transmission of avian influenza viruses between humans? (iii) What are the most effective interventions for reducing the probability that an influenza strain with pandemic potential will evolve? Our results suggest that if evolutionary emergence of past pandemics has occurred primarily through viral reassortment in humans, then thousands of avian influenza virus infections in humans must have occurred each year for the past 250 years. Analyses also show that if there is epidemiologically significant variation among avian influenza virus genotypes, then avian virus outbreaks stemming from repeated cross-species transmission events result in a greater likelihood of a pandemic strain evolving than those caused by low-level transmission between humans. Finally, public health interventions aimed at reducing the duration of avian virus infections in humans give the greatest reduction in the probability that a pandemic strain will evolve.
Keywords: viral reassortment, H5, H7, H9, avian influenza, emerging diseases
1. Introduction
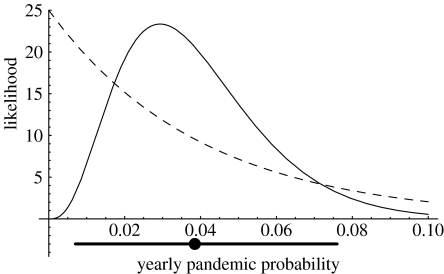
Influenza pandemics are characterized by the worldwide spread of novel influenza strains for which most of the population lacks substantial immunity (Webster et al. 1992; Cox & Subbarao 2000). These pandemic strains typically cause heightened morbidity and mortality (Cox & Subbarao 2000), and they tend to drive previous strains of the influenza virus to extinction (Cox & Subbarao 2000; Earn et al. 2002). Anecdotal evidence suggests that there have been approximately 10 influenza pandemics during the past 250 years (Webster 1998; Reid & Taubenberger 2003), indicating that the probability of a pandemic occurring in any given year has averaged about 4% over the past two-and-a-half centuries. A more reliable estimate of the average yearly probability of a pandemic is obtained by combining this anecdotal estimate with data on inter-pandemic intervals collected from the last century (Cox & Subbarao 2000; Reid et al. 2004). This yields a conservative 95% support interval for the yearly pandemic probability of 0.7–7.6%, with a point estimate of 3.9% (figure 1; see also the electronic supplementary material).
There are a variety of processes that might give rise to influenza strains with pandemic potential, and evaluating the significance of each of these has attracted much research effort. Molecular analyses suggest that the two previous pandemics involved reassortment between human-adapted and avian-adapted influenza viruses (Webster et al. 1992; Reid & Taubenberger 2003; Reid et al. 2004). The 1957 pandemic probably involved reassortment between an avian H2N2 influenza virus and a human H1N1 influenza virus (Scholtissek et al. 1978; Kawaoka et al. 1989; Schafer et al. 1993) and the 1968 pandemic probably involved reassortment between an avian H3Nx influenza virus and a human H2N2 influenza virus (Scholtissek et al. 1978; Bean et al. 1992). Even in these relatively well-documented cases, however, the species in which reassortment took place is not known.
It was initially suggested that viral reassortment most probably takes place in pigs because avian influenza viruses typically replicate poorly in humans (Beare & Webster 1991), whereas swine have epithelial receptors for both avian and human influenza (Ito et al. 1998). The recent cases of H5N1 (Claas et al. 1998; Shortridge et al. 1998; Subbarao et al. 1998) and H9N2 (Peiris et al. 1999) avian influenza in humans, however, clearly call this belief into question, and reassortment in humans is now regarded as a distinct possibility (Palese 2004; Ferguson et al. 2005; Horimoto & Kawaoka 2005; Longini et al. 2005).
Mathematical models can play a useful role in clarifying the potential significance of different routes of pandemic emergence, as well as in evaluating the most effective public health interventions for reducing the probability of pandemics. In this paper, we focus on a subset of potential routes for the emergence of pandemic strains, namely the evolutionary adaptation of avian influenza viruses in humans (via reassortment and/or mutation). We develop a stochastic model for these processes and use it to address three main questions: (i) what is the minimum annual number of avian influenza virus infections in humans that is required to explain the historical rate of pandemic emergence? The answer to this question provides some guidance to the levels of avian influenza virus infection in humans that we expect to observe if pandemic strains evolve via reassortment in humans; (ii) are such avian influenza infections in humans more likely to give rise to pandemic strains if they are driven by repeated cross-species introductions, or by low-level transmission of avian influenza viruses between humans? The answer to this question can provide some guidance to the conditions that are most likely to result in the evolution of pandemic strains; and (iii) what are the most effective public health interventions for reducing the probability that an influenza strain with pandemic potential will arise?
2. The model
We suppose that pandemic influenza strains arise via direct adaptation of avian influenza viruses to humans (i.e. there is no third species involved, such as swine). The generation of human-adapted strains during an avian influenza virus infection in humans might occur via two distinct processes: by reassortment between avian and human influenza viruses, or by mutation. Reassortment has received more attention in the literature, probably because the two previous pandemics have involved this process. Nevertheless, it is not yet known whether reassortment alone was sufficient to spawn these pandemics, or whether other evolutionary processes such as mutation were also required (Webster et al. 1992; Horimoto & Kawaoka 2005; Taubenberger et al. 2005). Furthermore, molecular analyses for the 1918 pandemic suggest that even evolution via mutation alone (and possibly recombination) might have been sufficient to generate the strain causing this pandemic (Webster et al. 1992; Gibbs et al. 2001a,b, 2002; Webby & Webster 2001; Fanning et al. 2002; Worobey et al. 2002; Reid & Taubenberger 2003; Reid et al. 2004; Webby et al. 2004; Horimoto & Kawaoka 2005; Oxford et al. 2005; Taubenberger et al. 2005). Consequently, it is important to include both reassortment and mutation in the model.
First consider reassortment in humans, and imagine a focal human infected with an avian influenza virus. There is some chance that this focal person will become concurrently infected with human influenza, potentially leading to co-infection within some cells. If co-infection occurs, then there is some chance of reassortment between the human and avian influenza viruses, resulting in a novel strain with pandemic potential. Furthermore, the probability of such a reassortant strain causing a pandemic will probably depend on the genotype of the avian influenza virus with which reassortment took place.
At the same time, the avian influenza virus infecting the focal individual is also subject to mutation as it replicates. Given that the focal human represents a novel host, we might expect some of these mutations to be selectively beneficial. If any of the mutations that appear result in a sufficient increase in its reproduction number, then this mutational process alone can give rise to a pandemic-capable strain as well.
Either reassortment or adaptation via mutation must occur for a single infected human to cause a pandemic, but this individual might also transmit the original avian influenza virus to other humans (Bridges et al. 2000, 2002; Ungchusak et al. 2005), and these secondary infections might initiate a pandemic via the same processes. Additionally, more than one individual might acquire an avian influenza infection directly from the avian source in any given year. Furthermore, there will typically be several avian influenza virus genotypes circulating in birds, each of which might differ in their capacity to give rise to a pandemic strain once transmitted to humans (Li et al. 2004; Lipatov et al. 2004; Webster & Hulse 2004; Wan et al. 2005).
All the above stochastic events can be combined within a single model using a continuous-time, multi-type branching process. We define as the expected number of cross-species transmission events per year and R0,i as the reproduction number of an avian influenza virus of genotype i in humans prior to any evolutionary change (this implies R0,i<1; Anderson & May 1991). Further, once a pandemic-capable strain has arisen (i.e. a strain with a reproduction number greater than 1), it might still be lost by chance; therefore, we define Pi as the probability that such a strain actually causes a pandemic (given that it arose from an avian influenza virus of genotype i). Standard results from branching processes then show that , where is the reproduction number of the pandemic-capable strain derived from avian virus genotype i.
With the above notation, an excellent approximation for the probability of a pandemic-capable strain arising and causing a pandemic in any given year is (see appendix A for a full derivation)
| 2.1a |
where
and E[ ] denotes the expectation taken over all avian virus genotypes i. The term is the probability that a single cross-species transmission event of avian influenza into humans will result in a pandemic-capable strain arising and causing a pandemic, averaged over all avian influenza genotypes. The quantity ai is (approximately; see below) the overall probability that a strain with pandemic potential emerges within any single avian influenza virus infection in humans. Equation (2.1a) can be intuited if we ignore genotypic variation among avian influenza viruses and suppose that a is very small, then (2.1a) can be further approximated as (Antia et al. 2003; André & Day 2005)
| 2.1b |
In equation (2.1b), 1/(1−R0) is the expected number of new infections generated by a single avian influenza virus infection in humans, each of which has a probability, aP, of undergoing adaptation and causing a pandemic. Given that there are such avian influenza introductions into humans, equation (2.1b) thereby gives the (approximate) overall probability of a pandemic occurring.
The above model demonstrates that the overall probability of pandemic emergence depends only on the mean probability of an avian influenza genotype giving rise to a pandemic (over all genotypes) and not on any other aspect of the distribution of genotypes. Furthermore, there are four fundamental parameters that appear: , R0,i, Pi and ai. The parameter ai is actually a composite parameter, which can be further broken down as ai=(ciri+μi)Li, where ci is the rate of co-infection with a human influenza virus; ri is the probability that reassortment between a human and an avian influenza virus occurs and gives rise to a strain with pandemic potential (given that co-infection of a cell has already happened); μi is the rate at which mutations conferring pandemic ability arise; and Li is the expected duration of an avian influenza virus infection in humans (all specific to avian influenza genotype i). Consequently, ai is the expected number of adaptation events (reassortment or mutation) that occur during a single avian influenza infection. However, if this is very small, then it can also be interpreted as the probability that adaptation occurs in any single avian influenza infection.
Equation (2.1a) reveals, not surprisingly, that, in principle, the accumulation of beneficial mutations alone (represented by the parameter μi) can be just as potent a factor in the emergence of pandemic strains as reassortment with human influenza viruses (represented by the product ciri). Both factors enter the composite parameter, ai, in the same way; therefore, which of these is most significant will depend on their relative magnitudes. The process of reassortment depends on the force of infection of human influenza, ci, as well as on the probability of reassortment into a pandemic strain once co-infection has occurred, ri. On the other hand, the process of adaptation via mutation is independent of the force of infection and depends only on the rate at which mutations conferring pandemic potential arise, μi. Thus, one interesting avenue of future empirical research would be to estimate the relative magnitude of ciri versus μi for different avian influenza genotypes.
3. Results
The model developed above can now be used to address the three questions posed in §1.
(a) What is the minimum annual number of avian influenza virus infections in humans required?
To simplify matters, we will suppose that there is very little variation among avian influenza genotypes, and approximate the mean probability that a single cross-species transmission event results in pandemic emergence (i.e. ) by evaluating it at the average values of its composite parameters, i.e.
| 3.1 |
with . However, it can be shown that variation in each of the parameters often tends to have opposing effects on the value of ; therefore, we might expect that this approximation holds under somewhat broader conditions (T. Day, J.-B. André and A. Park 2006, unpublished data). In what follows, we will simplify the notation by dropping the over-bars on each of these parameters.
With approximation (3.1), equation (2.1a) involves five quantities: the probability of a pandemic, Λ, and the four parameters , R0, P and a. Figure 1 presents the best available estimate of the annual probability of a pandemic, Λ, and our goal is to use this in equation (2.1a) to predict the minimum values of and R0 that would be required to yield this estimate (since together they determine the number of avian influenza infections in humans each year, via introductions and human–human transmission, respectively). Therefore, we need some estimate of the two remaining parameters, P and a.
Figure 1.
Results of a Bayesian analysis for the probability of a pandemic occurring in any given year. The dashed curve indicates prior probability density of the pandemic probability based on anecdotal information. The solid curve indicates posterior probability density of the pandemic probability once data from the past century are incorporated (see electronic supplementary material). The thick bar and dot below horizontal axis are the 95% support interval and the point estimate, respectively.
Because we are seeking minimum estimates, we can set P=1, as this will lead to an underestimate of the values of and R0 required. Furthermore, here we are interested solely in the process of reassortment in humans; therefore, we set μ=0. Finally, a rough point estimate for the product cL can be obtained from existing data (Ferguson et al. 2004; appendix A), leaving only the parameter, r. This is the probability that, given co-infection with human influenza, reassortment will occur between the avian and human influenza viruses and yield a strain with pandemic potential. Little is known about the value of r, but again, because we are seeking minimum estimates, we can make progress by exploring values of r with different orders of magnitude. The parameter r is almost certainly less than 1; therefore, we use the next two orders of magnitude, i.e. r=0.1 and 0.01. In reality, r is probably still much smaller; therefore, our calculations will again result in an underestimate of and R0.
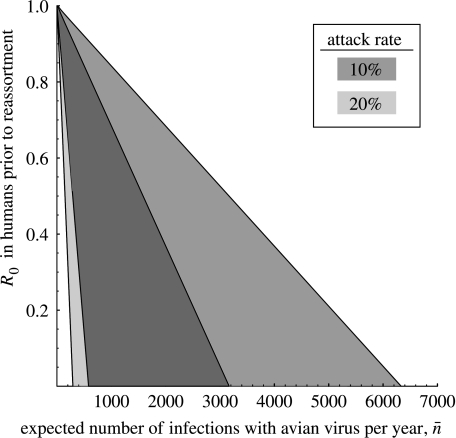
Figure 2 plots the range of values that R0 and must take in order to give rise to a probability of pandemic that lies within the 95% support interval Λ=0.7–7.6%. Hundreds to thousands of avian influenza virus infections must occur in the human population each year if viral reassortment in humans is to explain the historical occurrence of pandemics. This number of infections is certainly within the realm of possibility, but it is also worth remembering that allowing for pandemic strains to arise via the accumulation of beneficial mutations (i.e. μ≠0) can greatly relax these requirements. It is well appreciated that adaptation via mutation and/or recombination is likely to occur during the emergence of pandemic strains (Horimoto & Kawaoka 2005), but such adaptation alone has recently been suggested as a potential cause of at least one past pandemic as well (Taubenberger et al. 2005; but see Antonovics et al. 2006; Gibbs & Gibbs 2006; Taubenberger et al. 2006). Therefore, the significance of this additional route for the evolutionary emergence of pandemic strains should not be neglected.
Figure 2.
Shaded areas are combinations of and R0 required to generate a pandemic probability via reassortment in humans that falls within the 95% support interval presented in figure 1. Shades correspond to attack rates of human influenza (i.e. probability of an individual acquiring human influenza per year) and thus to different rates of co-infection, c. Results assume that r=0.01; if r=0.1 is assumed instead, the values on the horizontal axis simply need to be divided by 10.
Before proceeding further, it is worth noting that although there is considerable uncertainty involved in the parameter values used in figure 2, there are at least three reasons to believe that the above conclusions might be conservative. First, it was assumed that all individuals contracting both avian and human influenza have cells that are then co-infected with both, whereas some of these individuals will harbour the two viruses in different parts of their body (e.g. respiratory tract for human influenza versus superficial infections, such as conjunctivitis, for avian influenza), thereby precluding reassortment. Second, we assumed that once a pandemic-capable strain arises from reassortment within an individual, the infection that it causes in this index case is unaffected by the fact that this individual is also still co-infected with human and avian influenza. More realistically, such reassortants might often be hampered by cross-reaction with the immune response stimulated by the avian and human influenza viruses, thereby limiting their spread. Third, we assumed that once a pandemic strain arising from reassortment makes it out of the index case, it goes on to cause a pandemic with probability one. Typically, however, there is a reasonable chance that it will instead go extinct purely through stochasticity. For example, recent estimates place the reproduction number of the 1918 pandemic strain at approximately 3 (Mills et al. 2004), which gives a probability of loss through stochasticity of 0.33. Consequently, the overall probability of pandemic emergence would be reduced by a factor of approximately 2/3 (see equation (2.1b)).
(b) Cross-species introductions versus human–human transmission
The above results provide an estimate for the minimum number of avian influenza virus infections that must occur in humans each year to explain the historical rate of pandemic occurrence. These infections can arise as a result of two different processes: repeated cross-species introductions of avian influenza into the human population (the parameter in the model) and/or low levels of human–human transmission (the parameter R0 in the model; Bridges et al. 2000, 2002; Ferguson et al. 2004; Ungchusak et al. 2005). A natural question to ask then is, does it matter which of these routes gives rise to avian influenza infections in humans with respect to the probability of evolutionary emergence of pandemic influenza?
The simplest way to address this question is to suppose that there is a total of N avian influenza infections in humans in a given year, and to compare the probability of emergence in the case where all these infections result from separate cross-species introductions to that where they all result from a single cross-species introduction, followed by human–human transmission. If we use αi to denote the probability that any single infection with avian influenza genotype i results in the emergence of a pandemic strain from that individual (as opposed to πi, which is the probability of such a strain arising from that individual, or any of its ‘descendant’ infections), then a calculation analogous to that of equation (A 1) (see appendix A) gives the probability of emergence for each scenario as
| 3.2a |
and
| 3.2b |
where over-bars denote an expectation taken over all avian influenza genotypes.
Equations (3.2a) and (3.2b) differ only in how the expectation is taken and, as a result, Jensen's inequality implies that (3.2a) is larger than (3.2b). This indicates that the probability of emergence is largest for the case where the infections arise from separate introductions. For example, if there are two avian influenza genotypes, with ρ1=0.01, ρ2=0.99, α1=0.01 and α2=0, and if N=500, then the probability of emergence via separate introductions is nearly five times larger than that from human–human transmission (Λintroductions≈4.9% versus Λtransmission≈1%). If we view each infection as a trial in which emergence can occur, multiple introductions result in the trials being conducted with potentially new avian influenza virus genotypes each time, whereas human–human transmission results in the trials being conducted with the same avian influenza genotype repeatedly. As a result, the latter is less likely to yield the evolutionary emergence of a pandemic strain whenever there is significant epidemiological variability among genotypes of avian influenza.
(c) Effect of different public health interventions
Several recent mathematical analyses have focused on evaluating different potential interventions for controlling a pandemic once a pandemic strain has appeared (Ferguson et al. 2005; Longini et al. 2005), but relatively few studies have examined how such interventions might affect the initial evolutionary emergence of such strains in the first place. We now use the above probabilistic analyses to evaluate how different types of public health interventions affect the probability of evolutionary emergence (Webby & Webster 2003; Webster & Hulse 2004; Ferguson et al. 2005).
Possible public health interventions in the face of avian influenza infections in humans include limiting contact between humans and poultry (or other sources of foreign influenza viruses; Webster & Hulse 2004), employing vaccines specific to avian influenza viruses or prophylactic antiviral medication in high-risk groups (e.g. poultry workers; Lipatov et al. 2004; Webster & Hulse 2004; Horimoto & Kawaoka 2005; Longini et al. 2005), isolating individuals infected with avian influenza viruses, or administering therapeutic antiviral medication to help clear infections with avian influenza viruses (Lipatov et al. 2004; Horimoto & Kawaoka 2005; Longini et al. 2005). Any intervention is likely to affect more than one parameter of the model, but one can draw some rough correspondences (table 1), and then calculate the elasticity of the pandemic probability, Λ, to changes in each parameter (table 1).
Table 1.
Elasticity of pandemic probability to changes in each of the parameters (i.e. the percentage by which the pandemic probability decreases with a small percentage change in each parameter) listed in order of magnitude (figure 3). (The calculation is simplified by first supposing that both the rate of co-infection, c, and the rate of beneficial mutations arising, μ, are small. In this case, if we ignore genotypic variation among avian influenza strains, we can use equation (2.1b). The final column lists potential interventions that can affect the parameter in question.)
| parameter | interpretation | elasticity | potential intervention |
|---|---|---|---|
| L | duration of an avian virus infection in humans | (1/(1−R0)) | therapeutic antivirals |
| b | rate of production of avian virus infections in humans by humans (R0=bL) | (R0/(1−R0)) | isolation of humans with avian virus infections; therapeutic antivirals |
| P | probability of a pandemic, given that a strain with pandemic potential arises | 1 | vaccination, with vaccine specific to newly emerged pandemic strain |
| expected number of avian virus introductions into humans per year | 1 | reduced contact of humans with avian sources; vaccination specific to avian strains | |
| c | rate of co-infection with human influenza | (cr/(cr+μ)) | isolation of humans with avian virus infections |
| r | probability of reassortment, given that co-infection has occurred | (cr/(cr+μ)) | therapeutic antivirals |
| μ | rate of within-host adaptation through viral competition | (μ/(cr+μ)) | therapeutic antivirals |
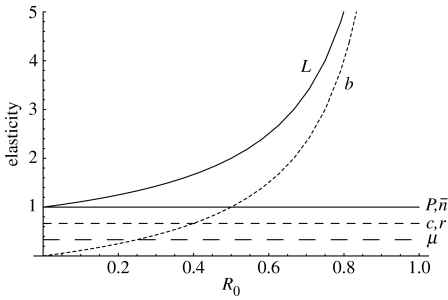
Regardless of both the mode by which pandemic strains arise (i.e. mutation versus reassortment) and the parameter values of the model, interventions that affect the duration of avian influenza virus infections in humans are predicted to provide the best overall reduction in the probability of a pandemic (figure 3; table 1). Interventions that reduce transmissibility of avian influenza among humans can also have a substantial effect, but only when the avian influenza reproduction number in humans is relatively large. Otherwise, this intervention has the smallest influence (figure 3). Finally, reductions in the number of cross-species introductions per year, or in the likelihood of a pandemic occurring once a pandemic-capable strain has arisen, tend to have intermediate effects (figure 3; table 1).
Figure 3.
Elasticity of the pandemic probability to changes in each of the model parameters (i.e. the percentage by which the pandemic probability decreases with a small percentage change in each parameter). Results are from table 1 and plotted as a function of the reproduction number of avian influenza in humans. The relative ordering of the line for μ and c,r is determined by the relative magnitude of these parameters (table 1), but both lines are always constant and less than 1.
4. Discussion
The first conclusion that can be drawn from the above probabilistic analyses is that, in the absence of extremely high rates of reassortment, a high level of avian influenza virus infection in humans is required every year for the past 250 years, if reassortment in humans has been the primary cause of past influenza pandemics. Although such levels have not been documented, it is nevertheless possible that such extensive infection occurs every year if the majority of these infections are indistinguishable from human influenza infections, or if they are asymptomatic. From the standpoint of the above calculations, the only requirement is that these avian influenza infections occur within tissues of the human body that are also typically infected by human influenza. Specifically, they would probably have to be respiratory avian influenza infections as opposed to superficial infections like conjunctivitis.
If such high levels of avian influenza infection have not occurred in humans, then the only way for reassortment in humans to explain the past frequency of pandemics is if r, the probability of reassortment into a pandemic strain with a large reproductive number, is very high. Specifically, values of r of the order of 50–100% would be required. Although this seems unrealistically high, there is not yet sufficient data available to rule out this possibility. In any event, taken together, the above two requirements suggest another two very fruitful avenues for future empirical investigation: estimating the yearly level of avian influenza virus infection in humans (e.g. via serological studies; Meijer et al. 2006), and estimating r (e.g. via experimental co-infection studies in model organisms). With such estimates, equation (2.1a), along with the estimate of figure 1, can then be used to develop more precise conclusions about the importance of reassortment in humans.
The probabilistic analyses used here also show that adaptation via mutation can be a powerful additional factor that increases the likelihood of evolutionary emergence of strains with pandemic potential. It is well appreciated that adaptation via the accumulation of beneficial mutations is likely to occur during the emergence of pandemic influenza, as natural selection ‘fine tunes’ a newly emerging influenza virus to humans (Horimoto & Kawaoka 2005). Recently, however, empirical evidence has suggested that even such adaptation might have been sufficient to cause the 1918 influenza pandemic (Taubenberger et al. 2005). Although this conclusion has been called into question (Antonovics et al. 2006; Gibbs & Gibbs 2006; Taubenberger et al. 2006), the above analyses lend theoretical support to this idea and raise the possibility that this mode of pandemic emergence might have played a significant role in other pandemics throughout history as well.
Although there are some reasons to believe that the conclusions drawn above about the level of avian influenza infection in humans are conservative, our analysis employs the questionable assumption that the probability of a pandemic has been constant throughout history. More realistically, the probability of a pandemic probably varies from year to year, and perhaps has also changed systematically over time (Webby & Webster 2003; Lipatov et al. 2004). Unfortunately, very little data are available to conduct any meaningful analysis of this question. Furthermore, it is not even qualitatively clear from the anecdotal evidence whether the probability of emergence has increased recently (e.g. owing to large-scale poultry farms) or whether it has decreased (e.g. because people are now less apt to live in close quarters with poultry). Consequently, the results presented here are best viewed as historical averages.
Regardless of whether reassortment or mutation generates a pandemic strain, it is also interesting to speculate on how we might expect the initial emerging pandemic strain to have adapted to humans. Given that this new strain initially arises in a human host who also contains the original avian influenza virus, two different modes of adaptation might be expected depending on the characteristics of original avian influenza infection. For example, if avian influenza viruses tend to induce very mild infections in humans, then pandemic-capable strains that emerge might gain their advantage through either an evolved increase in transmissibility or an increase in infection duration (e.g. through immune evasion). On the other hand, if avian influenza viruses tend to induce high levels of mortality in humans (e.g. H5N1), then we might expect emerging strains to have evolved largely by having a higher transmissibility. Increased infection duration would be of no initial selective advantage in this case because it would be the co-infecting avian influenza virus that determines the lifespan of the initial infection.
In addition to evaluating hypotheses for the evolutionary emergence of pandemic influenza, we have also used probabilistic analyses to determine the types of avian influenza virus outbreaks in humans that are most likely to result in the evolution of a strain with pandemic potential. Results show that multiple introductions of avian influenza viruses into humans each year result in a greater likelihood of evolutionary emergence than very few introductions coupled with low levels of human–human transmission. Multiple introductions result in a greater sampling of avian influenza virus genotypes from the avian source population, and this thereby increases the odds of a genotype that yields a highly transmissible strain upon reassortment being introduced into the human population.
However, these results do not imply that human–human transmission of avian influenza is less worrisome than multiple introductions. Indeed, outbreaks of avian influenza stemming from human–human transmission might indicate that a pandemic strain has already evolved and is spreading. Rather, our results reveal that, during the initial process of the evolution of such a strain, if avian influenza outbreaks in humans tend to arise from multiple introductions as opposed to few introductions coupled with human–human transmission, then the likelihood of such evolution occurring is expected to be higher.
Finally, the probabilistic analyses used in this paper reveal that public health interventions affecting the duration of avian influenza virus infections in humans tend to provide the best overall reduction in the probability of a pandemic, regardless of whether pandemics arise from reassortment or from adaptation via mutation. Such changes impart three benefits: they reduce the total number of infections in humans; they reduce the likelihood of co-infection by reducing the duration of avian influenza virus infections; and they reduce the time during which beneficial mutations might arise. It is also possible, however, that if the reproduction number of avian influenza in humans is relatively large, then interventions that reduce the transmission rate are also very effective (figure 3).
Taken together, these results suggest that interventions such as therapeutic antiviral medication are expected to be one of the best options for reducing the likelihood that a pandemic influenza strain will evolve. Such interventions will reduce both the duration of avian influenza infections in humans and the transmission rate. Furthermore, given that therapeutic antiviral medication is likely to be a powerful tool for controlling the spread of pandemic influenza once it has evolved (Ferguson et al. 2003, 2005; Longini et al. 2005), this suggests that such antivirals might provide a very significant, double benefit: one being evolutionary, and the other being epidemiological. However, it should also be noted that applying such treatment to high-risk groups as a prophylaxis might also be a valuable approach, since the benefits of antiviral therapy will be best realized if the lag between the start of infection and the use of antivirals is as small as possible. All these assume, of course, that such antiviral drugs can produce a significant reduction in the duration of avian influenza virus infection in humans.
Throughout our analysis we have focused on a subset of potential processes that might be involved in the evolutionary emergence of pandemic influenza (reassortment and/or mutational changes during infection in humans). Another possibility, however, is that reassortment is the primary engine of influenza pandemics, but that it occurs in a third host species, such as swine. Although we have not examined this hypothesis explicitly, our analysis, nevertheless, sheds some light on this possibility.
The process of reassortment in swine is conceptually identical to that of reassortment in humans, except for two factors. First, any newly reassorted strain in pigs must make its way back in to humans before it can cause a pandemic, and this extra step in the process reduces the likelihood of emergence as compared with reassortment in humans. On the other hand, because each of the events leading to reassortment (e.g. co-infection, transmission among hosts, etc.) might occur more readily in pigs than it does in humans (e.g. pigs can more readily be infected with both avian and human influenza; Beare & Webster 1991), this can increase the probability of emergence. Unfortunately, because these two factors counteract one another, it is not possible to determine whether the pandemic probability via reassortment in pigs is expected to be larger or smaller than that from reassortment in humans. In any case, however, if reassortment in pigs is a primary source of pandemic strains, then our analysis does suggest that very high levels of avian influenza infection in swine are then expected (just as it is in humans).
Finally, it should also be noted that our analyses of reassortment assume that the compounding of events giving rise to the evolutionary emergence of a pandemic strain occurs within a single year. Alternatively, the evolutionary steps involved might build upon one another over a series of years. For example, a foreign influenza virus lineage (e.g. avian) might become permanently established in swine in one year, and then circulate for several years in a self-sustaining way (as has been documented; Olsen 2002). Co-infection and reassortment could then occur in some later year (Peiris et al. 2001). It is more difficult to model this scenario precisely because the probability of a pandemic is then no longer constant from year to year. Nevertheless, this is yet another possibility.
Acknowledgments
We thank D. Earn, C. Fraser, S. Gandon, B. Grenfell, J. Plotkin, A. Read, M. Woolhouse and two anonymous reviewers for their helpful comments and/or discussions. This research was supported by grants to T.D. from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program. A.P. was supported by a grant from the Mathematics of Information Technology and Complex Systems and J.B.A. by a grant from Queen's University.
Appendix A
Consider that a single human is infected with an avian influenza virus of genotype i, and use πi to denote the probability that this ultimately leads to a pandemic-capable strain emerging and causing a pandemic. This probability includes the possibility that the initially infected individual itself gives rise to a pandemic-capable strain, as well as the possibility that any of the ‘descendant’ infections generated by this initially infected individual gives rise to a pandemic-capable strain.
We suppose that there are σ genotypes of avian influenza, and that the number of cross-species transmission events occurring in any given year is a random variable, N. The probability of a pandemic occurring in any given year is then
| A1a |
where P(N=n) is the probability density of N; ρi is the frequency of avian influenza genotype i in the bird population; and k1, k2, … , kσ denote the (random) number of introduction of each genotype (with k1+k2+ ⋯ +kσ=n). The factor is the probability that all n introduced avian influenza virus strains fail to generate a pandemic, while is the (multinomial) probability that, if a total of n strains are introduced, k1 will be of genotype 1, k2 will be of genotype 2, etc.
Equation (A 1a) can be simplified using the multinomial theorem to obtain
| A1b |
where is the probability of an avian influenza strain giving rise to a pandemic in humans, averaged over all avian influenza genotypes. Cross-species transmission is a relatively rare event; therefore, the probability density P(N=n) is taken to be a Poisson distribution (i.e. , where is the mean number of events per year). In this case, (A 1b) simplifies to
| A1c |
Finally, under the assumption that is not too large, (A 1c) can be approximated as
| A1d |
Equation (A 1d) is an excellent approximation whenever is not greater than approximately 0.1, which is always the case for realistic parameter values.
To complete the analysis, πi must be calculated for each avian influenza genotype. We use a continuous-time, two-type branching process with constant rates (Jagers 1975; Antia et al. 2003; André & Day 2005). In reality, the rates in question (see below) will probably vary during the course of an infection, but too little data are currently available to warrant using such a complex model. The two ‘types’ of entities modelled are: (i) humans infected with an avian influenza virus and (ii) humans infected with a strain that is either a reassortment of an avian and a human influenza virus (and thus has pandemic potential) or a strain that has a new mutation conferring it with the ability to cause a pandemic. Here, we define the following (specific to an avian influenza virus of genotype i):
bi: ‘birth’ rate of new avian influenza infections in humans by an avian influenza infection in humans;
di: death plus recovery rate of an avian influenza infection in a human;
R0,i: reproduction number in human population of an avian influenza virus, R0,i=bi/di. This represents the average number of new avian influenza virus infections (in humans) that are produced via human–human transmission by a single avian influenza virus infection in humans (i.e. the actual number of transmission events produced by any given infection is a random variable with mean R0,i). R0,i is less than 1 and might even be zero because avian influenza viruses are not adapted to humans;
ci: rate of co-infection of cells of an individual that is infected with an avian influenza virus by human influenza. This depends on the force of infection of human influenza;
ri: probability of reassortment occurring, given that an individual is co-infected with a human and an avian influenza virus. For any avian influenza virus genotype, there are many different possible reassortant viruses depending upon the combination of gene segments that are combined, but we assume that the most transmissible combination is always produced, given that reassortment occurs and that this combination has pandemic potential;
Li: average duration of an avian influenza infection in humans, Li=1/di;
: reproduction number in human population of a virus that is a reassortment of a human influenza virus and an avian influenza virus of genotype i;
Pi: probability that if a reassortant virus with reproduction number arises within a human, it results in a pandemic (as opposed to dying out by chance). for reassortant virus with , otherwise it is zero.
With this notation, the appendix of André & Day (2005) can be followed to obtain
| A2 |
which then yields equation (2.1a).
Parameter estimates. An estimate for the probability of a secondary infection with human influenza, cL, can be derived as follows (Ferguson et al. 2004). Assume that the attack rate of human influenza is 1/5–1/10, and that the typical influenza season lasts 12 weeks or 84 days. This implies that any given individual has a probability of approximately 1/(5×84)−1/(10×84) of being infected with human influenza each day during the influenza season. If we suppose that only 1 day during an avian infection in humans is open to the possibility of co-infection with human influenza (Ferguson et al. 2004), then these are also estimates of the probabilities of co-infection, cL. Thus, we have cL=0.0012–0.0024. These calculations assume that the rate of co-infection is constant during the influenza season, but more generally the value of cL can be thought of as a yearly average probability of co-infection.
Supplementary Material
Bayesian estimate of yearly pandemic probability.
References
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- André J.-B, Day T. The effect of disease life history on the evolutionary emergence of novel pathogens. Proc. R. Soc. B. 2005;272:1949–1956. doi: 10.1098/rspb.2005.3170. doi:10.1098/rspb.2005.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antia R, Regoes R.R, Koella J.C, Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. doi:10.1038/nature02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics J, Hood M.E, Baker C.H. Was the 1918 flu avian in origin? Nature. 2006;440:E9. doi: 10.1038/nature04824. doi:10.1038/nature04824 [DOI] [PubMed] [Google Scholar]
- Bean W.J, Schell M, Katz J, Kawaoka Y, Naeve C, Gorman O, Webster R.G. Evolution of The H3 influenza-virus hemagglutinin from human and nonhuman hosts. J. Virol. 1992;66:1129–1138. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare A.S, Webster R.G. Replication of avian influenza viruses in humans. Arch. Virol. 1991;119:37–42. doi: 10.1007/BF01314321. doi:10.1007/BF01314321 [DOI] [PubMed] [Google Scholar]
- Bridges C.B, et al. Risk of influenza A (H5N1) infection among health care workers exposed to patients with influenza A (H5N1), Hong Kong. J. Infect. Dis. 2000;181:344–348. doi: 10.1086/315213. doi:10.1086/315213 [DOI] [PubMed] [Google Scholar]
- Bridges C.B, et al. Risk of influenza A (H5N1) infection among poultry workers, Hong Kong, 1997–1998. J. Infect. Dis. 2002;185:1005–1010. doi: 10.1086/340044. doi:10.1086/340044 [DOI] [PubMed] [Google Scholar]
- Claas E.C.J, Osterhaus A, van Beek R, De Jong J.C, Rimmelzwaan G.F, Senne D.A, Krauss S, Shortridge K.F, Webster R.G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. doi:10.1016/S0140-6736(97)11212-0 [DOI] [PubMed] [Google Scholar]
- Cox N.J, Subbarao K. Global epidemiology of influenza: past and present. Ann. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. doi:10.1146/annurev.med.51.1.407 [DOI] [PubMed] [Google Scholar]
- Earn D.J.D, Dushoff J, Levin S.A. Ecology and evolution of the flu. Trends Ecol. Evol. 2002;17:334–340. doi:10.1016/S0169-5347(02)02502-8 [Google Scholar]
- Fanning T.G, Slemons R.D, Reid A.H, Janczewski T.A, Dean J, Taubenberger J.K. 1917 avian influenza virus sequences suggest that the 1918 pandemic virus did not acquire its hemagglutinin directly from birds. J. Virol. 2002;76:7860–7862. doi: 10.1128/JVI.76.15.7860-7862.2002. doi:10.1128/JVI.76.15.7860-7862.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N.M, Mallett S, Jackson H, Roberts N, Ward P. A population-dynamic model for evaluating the potential spread of drug-resistant influenza virus infections during community-based use of antivirals. J. Antimicrob. Chemother. 2003;51:977–990. doi: 10.1093/jac/dkg136. doi:10.1093/jac/dkg136 [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Fraser C, Donnelly C.A, Ghani A.C, Anderson R.M. Public health risk from the avian H5N1 influenza epidemic. Science. 2004;304:968–969. doi: 10.1126/science.1096898. doi:10.1126/science.1096898 [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Cummings D.A.T, Cauchemez S, Fraser C, Riley S, Meeyai A, Iamsirithaworn S, Burke D.S. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. doi:10.1038/nature04017 [DOI] [PubMed] [Google Scholar]
- Gibbs M.J, Gibbs A.J. Was the 1918 pandemic caused by a bird flu? Nature. 2006;440:E8. doi: 10.1038/nature04823. doi:10.1038/nature04823 [DOI] [PubMed] [Google Scholar]
- Gibbs M.J, Armstrong J.S, Gibbs A.J. Recombination in the hemagglutinin gene of the 1918 ‘Spanish flu’. Science. 2001a;293:1842–1845. doi: 10.1126/science.1061662. doi:10.1126/science.1061662 [DOI] [PubMed] [Google Scholar]
- Gibbs M.J, Armstrong J.S, Gibbs A.J. The haemagglutinin gene, but not the neuraminidase gene, of ‘Spanish flu’ was a recombinant. Phil. Trans. R. Soc. B. 2001b;356:1845–1855. doi: 10.1098/rstb.2001.0998. doi:10.1098/rstb.2001.0998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M.J, Armstrong J.S, Gibbs A.J. Questioning the evidence for genetic recombination in the 1918 “Spanish flu” virus—response. Science. 2002:296. doi: 10.1126/science.296.5566.211a. [DOI] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. doi:10.1038/nrmicro1208 [DOI] [PubMed] [Google Scholar]
- Ito T, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagers P. Wiley; New York, NY: 1975. Branching processes with biological applications. [Google Scholar]
- Kawaoka Y, Krauss S, Webster R.G. Avian-to-human transmission Of the Pb1 gene of influenza-A viruses in the 1957 and 1968 pandemics. J. Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K.S, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. doi:10.1038/nature02746 [DOI] [PubMed] [Google Scholar]
- Lipatov A.S, Govorkova E.A, Webby R.J, Ozaki H, Peiris M, Guan Y, Poon L, Webster R.G. Influenza: emergence and control. J. Virol. 2004;78:8951–8959. doi: 10.1128/JVI.78.17.8951-8959.2004. doi:10.1128/JVI.78.17.8951-8959.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longini I.M, Nizam A, Xu S.F, Ungchusak K, Hanshaoworakul W, Cummings D.A.T, Halloran M.E. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. doi:10.1126/science.1115717 [DOI] [PubMed] [Google Scholar]
- Meijer A, Bosman A, van de Kamp E, Wilbrink B, Holle M.D.R, Koopmans M. Measurement of antibodies to avian influenza virus A(H7N7) in humans by hemagglutination inhibition test. J. Virol. Methods. 2006;132:113–120. doi: 10.1016/j.jviromet.2005.10.001. doi:10.1016/j.jviromet.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Mills C.E, Robins J.M, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. doi:10.1038/nature03063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210. doi: 10.1016/s0168-1702(02)00027-8. doi:10.1016/S0168-1702(02)00027-8 [DOI] [PubMed] [Google Scholar]
- Oxford J.S, Lambkin R, Sefton A, Daniels R, Elliot A, Brown R, Gill D. A hypothesis: the conjunction of soldiers, gas, pigs, ducks, geese and horses in Northern France during the Great War provided the conditions for the emergence of the “Spanish” influenza pandemic of 1918–1919. Vaccine. 2005;23:940–945. doi: 10.1016/j.vaccine.2004.06.035. doi:10.1016/j.vaccine.2004.06.035 [DOI] [PubMed] [Google Scholar]
- Palese P. Influenza: old and new threats. Nat. Med. 2004;10:S82–S87. doi: 10.1038/nm1141. doi:10.1038/nm1141 [DOI] [PubMed] [Google Scholar]
- Peiris M, Yuen K.Y, Leung C.W, Chan K.H, Ip P.L.S, Lai R.W.M, Orr W.K, Shortridge K.F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. doi:10.1016/S0140-6736(99)03311-5 [DOI] [PubMed] [Google Scholar]
- Peiris J.S.M, Guan Y, Markwell D, Ghose P, Webster R.G, Shortridge K.F. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. doi:10.1128/JVI.75.20.9679-9686.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A.H, Taubenberger J.K. The origin of the 1918 pandemic influenza virus: a continuing enigma. J. Gen. Virol. 2003;84:2285–2292. doi: 10.1099/vir.0.19302-0. doi:10.1099/vir.0.19302-0 [DOI] [PubMed] [Google Scholar]
- Reid A.H, Taubenberger J.K, Fanning T.G. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nat. Rev. Microbiol. 2004;2:909–914. doi: 10.1038/nrmicro1027. doi:10.1038/nrmicro1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J.R, Kawaoka Y, Bean W.J, Suss J, Senne D, Webster R.G. Origin of the pandemic 1957 H2 influenza-A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. doi:10.1006/viro.1993.1319 [DOI] [PubMed] [Google Scholar]
- Scholtissek C, Rohde W, Vonhoyningen V, Rott R. Origin of human influenza-virus subtypes H2n2 and H3n2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. doi:10.1016/0042-6822(78)90153-8 [DOI] [PubMed] [Google Scholar]
- Shortridge K.F, et al. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. doi:10.1006/viro.1998.9488 [DOI] [PubMed] [Google Scholar]
- Subbarao K, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. doi:10.1126/science.279.5349.393 [DOI] [PubMed] [Google Scholar]
- Taubenberger J.K, Reid A.H, Lourens R.M, Wang R.L, Jin G, Fanning T.G. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. doi:10.1038/nature04230 [DOI] [PubMed] [Google Scholar]
- Taubenberger J.K, Reid A.H, Lourens R.M, Wang R, Jin G, Fanning T.G. Molecular virology: was the 1918 pandemic caused by a bird flu? Was the 1918 flu avian in origin? (Reply) Nature. 2006;440:E9–E10. doi:10.1038/nature04825 [Google Scholar]
- Ungchusak K, et al. Probable person-to-person transmission of avian influenza A (H5N1) New Engl. J. Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. doi:10.1056/NEJMoa044021 [DOI] [PubMed] [Google Scholar]
- Wan X.F, et al. Genetic characterization of H5N1 avian influenza viruses isolated in southern China during the 2003–04 avian influenza outbreaks. Arch. Virol. 2005;150:1257–1266. doi: 10.1007/s00705-004-0474-9. doi:10.1007/s00705-004-0474-9 [DOI] [PubMed] [Google Scholar]
- Webby R.J, Webster R.G. Emergence of influenza A viruses. Phil. Trans. R. Soc. B. 2001;356:1817–1828. doi: 10.1098/rstb.2001.0997. doi:10.1098/rstb.2001.0997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby R.J, Webster R.G. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. doi:10.1126/science.1090350 [DOI] [PubMed] [Google Scholar]
- Webby R, Hoffmann E, Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat. Med. 2004;10:S77–S81. doi: 10.1038/nm1151. doi:10.1038/nm1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G. Influenza: an emerging disease. Emerg. Infect. Dis. 1998;4:436–441. doi: 10.3201/eid0403.980325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G, Hulse D.J. Microbial adaptation and change: avian influenza. Revue Scientifique Et Technique De L' Office International Des Epizooties. 2004;23:453–465. doi: 10.20506/rst.23.2.1493. [DOI] [PubMed] [Google Scholar]
- Webster R.G, Bean W.J, Gorman O.T, Chambers T.M, Kawaoka Y. Evolution and ecology of influenza-A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Rambaut A, Pybus O.G, Robertson D.L. Questioning the evidence for genetic recombination in the 1918 “Spanish flu” virus. Science. 2002;296:211. doi: 10.1126/science.296.5566.211a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian estimate of yearly pandemic probability.